bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1645_orf3
Length=137
Score E
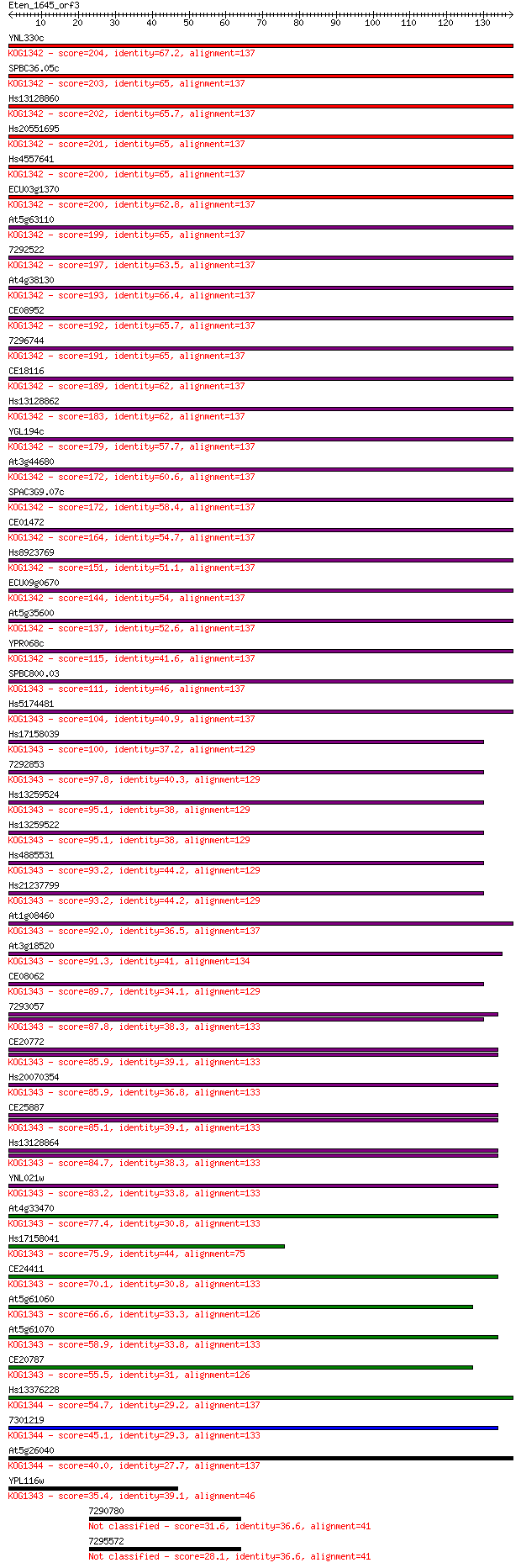
Sequences producing significant alignments: (Bits) Value
YNL330c 204 4e-53
SPBC36.05c 203 9e-53
Hs13128860 202 2e-52
Hs20551695 201 3e-52
Hs4557641 200 7e-52
ECU03g1370 200 7e-52
At5g63110 199 1e-51
7292522 197 4e-51
At4g38130 193 6e-50
CE08952 192 2e-49
7296744 191 2e-49
CE18116 189 1e-48
Hs13128862 183 1e-46
YGL194c 179 1e-45
At3g44680 172 2e-43
SPAC3G9.07c 172 2e-43
CE01472 164 3e-41
Hs8923769 151 4e-37
ECU09g0670 144 3e-35
At5g35600 137 6e-33
YPR068c 115 3e-26
SPBC800.03 111 3e-25
Hs5174481 104 5e-23
Hs17158039 100 7e-22
7292853 97.8 6e-21
Hs13259524 95.1 4e-20
Hs13259522 95.1 4e-20
Hs4885531 93.2 1e-19
Hs21237799 93.2 1e-19
At1g08460 92.0 3e-19
At3g18520 91.3 5e-19
CE08062 89.7 2e-18
7293057 87.8 6e-18
CE20772 85.9 2e-17
Hs20070354 85.9 2e-17
CE25887 85.1 3e-17
Hs13128864 84.7 4e-17
YNL021w 83.2 1e-16
At4g33470 77.4 8e-15
Hs17158041 75.9 2e-14
CE24411 70.1 1e-12
At5g61060 66.6 1e-11
At5g61070 58.9 2e-09
CE20787 55.5 3e-08
Hs13376228 54.7 5e-08
7301219 45.1 4e-05
At5g26040 40.0 0.001
YPL116w 35.4 0.033
7290780 31.6 0.43
7295572 28.1 4.9
> YNL330c
Length=433
Score = 204 bits (519), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 92/137 (67%), Positives = 110/137 (80%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCY+ND VL +E LR+ RVLY+D+D+HHGDGVEEAFYT+ RV+ CSFH
Sbjct 150 HHAKKSEASGFCYLNDIVLGIIELLRYHPRVLYIDIDVHHGDGVEEAFYTTDRVMTCSFH 209
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG FFPGTG L DIG+ G Y+VNVPL +GIDD Y S+F+ V+ +IME Y+P AVV
Sbjct 210 KYGEFFPGTGELRDIGVGAGKNYAVNVPLRDGIDDATYRSVFEPVIKKIMEWYQPSAVVL 269
Query 121 QCGADSVAGDRLGCFNL 137
QCG DS++GDRLGCFNL
Sbjct 270 QCGGDSLSGDRLGCFNL 286
> SPBC36.05c
Length=405
Score = 203 bits (516), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 111/137 (81%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND LAALE L++ RVLY+D+D+HHGDGVEE FYT+ RV+ CSFH
Sbjct 137 HHAKKREASGFCYVNDIALAALELLKYHQRVLYIDIDVHHGDGVEEFFYTTDRVMTCSFH 196
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
K+G +FPGTG ++D G+ G Y+VNVPL +GIDD+ Y S+F+ V+ IM+ +RPEAV+
Sbjct 197 KFGEYFPGTGHIKDTGIGTGKNYAVNVPLRDGIDDESYESVFKPVISHIMQWFRPEAVIL 256
Query 121 QCGADSVAGDRLGCFNL 137
QCG DS+AGDRLGCFNL
Sbjct 257 QCGTDSLAGDRLGCFNL 273
> Hs13128860
Length=482
Score = 202 bits (514), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/137 (65%), Positives = 112/137 (81%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VLA LE L++ RVLY+D+DIHHGDGVEEAFYT+ RV+ SFH
Sbjct 140 HHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFH 199
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG +FPGTG L DIG +G Y+VN PL +GIDD+ Y ++F+ VM ++ME+++P AVV
Sbjct 200 KYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAIFKPVMSKVMEMFQPSAVVL 259
Query 121 QCGADSVAGDRLGCFNL 137
QCG+DS++GDRLGCFNL
Sbjct 260 QCGSDSLSGDRLGCFNL 276
> Hs20551695
Length=488
Score = 201 bits (512), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 111/137 (81%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VLA LE L++ RVLY+D+DIHHGDGVEEAFYT+ RV+ SFH
Sbjct 141 HHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFH 200
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG +FPGTG L DIG +G Y+VN P+ +GIDD+ Y +F+ ++ ++ME+Y+P AVV
Sbjct 201 KYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQIFKPIISKVMEMYQPSAVVL 260
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS++GDRLGCFNL
Sbjct 261 QCGADSLSGDRLGCFNL 277
> Hs4557641
Length=488
Score = 200 bits (508), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 112/137 (81%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK+EASGFCYVND VLA LE L++ RVLY+D+DIHHGDGVEEAFYT+ RV+ SFH
Sbjct 141 HHAKKYEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHGDGVEEAFYTTDRVMTVSFH 200
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG +FPGTG L DIG +G Y+VN P+ +GIDD+ Y +F+ ++ ++ME+Y+P AVV
Sbjct 201 KYGEYFPGTGDLRDIGAGKGKYYAVNFPMCDGIDDESYGQIFKPIISKVMEMYQPSAVVL 260
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS++GDRLGCFNL
Sbjct 261 QCGADSLSGDRLGCFNL 277
> ECU03g1370
Length=416
Score = 200 bits (508), Expect = 7e-52, Method: Composition-based stats.
Identities = 86/137 (62%), Positives = 111/137 (81%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH K+ EASGFCYVND VL LE L++ R+LY+D+DIHHGDGVEEAFYT+ RV+ SFH
Sbjct 135 HHAKRGEASGFCYVNDIVLGILELLKYNKRILYIDIDIHHGDGVEEAFYTTDRVMTVSFH 194
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG +FPGTG + D GL +G GYSVNVPL +GIDD+ Y S+F+ ++ +++E Y+P A+V
Sbjct 195 KYGDYFPGTGMVTDTGLAKGKGYSVNVPLKDGIDDESYFSIFKPIVSKVIETYQPNAIVL 254
Query 121 QCGADSVAGDRLGCFNL 137
Q GADS++GD+LGCFNL
Sbjct 255 QSGADSLSGDKLGCFNL 271
> At5g63110
Length=471
Score = 199 bits (507), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 114/137 (83%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VL LE L+ RVLY+D+D+HHGDGVEEAFYT+ RV+ SFH
Sbjct 152 HHAKKSEASGFCYVNDIVLGILELLKMFKRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFH 211
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
K+G FFPGTG + D+G E+G Y++NVPL++G+DD+ + SLF+ ++ ++MEVY+PEAVV
Sbjct 212 KFGDFFPGTGHIRDVGAEKGKYYALNVPLNDGMDDESFRSLFRPLIQKVMEVYQPEAVVL 271
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS++GDRLGCFNL
Sbjct 272 QCGADSLSGDRLGCFNL 288
> 7292522
Length=521
Score = 197 bits (502), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 87/137 (63%), Positives = 108/137 (78%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VL LE L++ RVLY+D+D+HHGDGVEEAFYT+ RV+ SFH
Sbjct 138 HHAKKSEASGFCYVNDIVLGILELLKYHQRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFH 197
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG +FPGTG L DIG +G Y+VN+PL +G+DD Y S+F ++ ++ME ++P AVV
Sbjct 198 KYGEYFPGTGDLRDIGAGKGKYYAVNIPLRDGMDDDAYESIFVPIISKVMETFQPAAVVL 257
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS+ GDRLGCFNL
Sbjct 258 QCGADSLTGDRLGCFNL 274
> At4g38130
Length=501
Score = 193 bits (491), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 91/137 (66%), Positives = 110/137 (80%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VLA LE L+ RVLYVD+DIHHGDGVEEAFY + RV+ SFH
Sbjct 148 HHAKKCEASGFCYVNDIVLAILELLKQHERVLYVDIDIHHGDGVEEAFYATDRVMTVSFH 207
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
K+G +FPGTG ++DIG G YS+NVPL +GIDD+ Y LF+ +M ++ME++RP AVV
Sbjct 208 KFGDYFPGTGHIQDIGYGSGKYYSLNVPLDDGIDDESYHLLFKPIMGKVMEIFRPGAVVL 267
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS++GDRLGCFNL
Sbjct 268 QCGADSLSGDRLGCFNL 284
> CE08952
Length=461
Score = 192 bits (487), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 90/137 (65%), Positives = 106/137 (77%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCY ND VL LE L++ RVLYVD+D+HHGDGVEEAFYT+ RV+ SFH
Sbjct 144 HHAKKSEASGFCYTNDIVLGILELLKYHKRVLYVDIDVHHGDGVEEAFYTTDRVMTVSFH 203
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
KYG FFPGTG L+DIG +G YSVNVPL +GI D Y S+F+ +M ++ME + P AVV
Sbjct 204 KYGDFFPGTGDLKDIGAGKGKLYSVNVPLRDGITDVSYQSIFKPIMTKVMERFDPCAVVL 263
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS+ GDRLG FNL
Sbjct 264 QCGADSLNGDRLGPFNL 280
> 7296744
Length=438
Score = 191 bits (486), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 89/138 (64%), Positives = 109/138 (78%), Gaps = 1/138 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND V+ LE L++ RVLY+D+D+HHGDGV+EAFY + RV+ SFH
Sbjct 139 HHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDVHHGDGVQEAFYLTDRVMTASFH 198
Query 61 KYG-HFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
KYG +FFPGTG + +IG E G YSVNVPL EGIDDQ Y +F+ ++ IM+ YRP A+V
Sbjct 199 KYGNYFFPGTGDMYEIGAESGRYYSVNVPLKEGIDDQSYFQVFKPIISAIMDFYRPTAIV 258
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+AGDRLGCF+L
Sbjct 259 LQCGADSLAGDRLGCFSL 276
> CE18116
Length=465
Score = 189 bits (480), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 85/137 (62%), Positives = 107/137 (78%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCY ND VLA LE L+ RVLY+D+D+HHGDGVEEAFYT+ RV+ SFH
Sbjct 139 HHAKKSEASGFCYSNDIVLAILELLKHHKRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFH 198
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
K+G +FPGTG L+D+G G Y++NVPL +G+DD Y +F+ +M ++M ++PEAVV
Sbjct 199 KHGEYFPGTGDLKDVGAGSGKYYALNVPLRDGVDDVTYERIFRTIMGEVMARFQPEAVVL 258
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS+AGDRLG FNL
Sbjct 259 QCGADSLAGDRLGVFNL 275
> Hs13128862
Length=428
Score = 183 bits (464), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 85/138 (61%), Positives = 107/138 (77%), Gaps = 1/138 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND V+ LE L++ RVLY+D+DIHHGDGV+EAFY + RV+ SFH
Sbjct 134 HHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEAFYLTDRVMTVSFH 193
Query 61 KYG-HFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
KYG +FFPGTG + ++G E G Y +NVPL +GIDDQ Y LFQ V++Q+++ Y+P +V
Sbjct 194 KYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVINQVVDFYQPTCIV 253
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+ DRLGCFNL
Sbjct 254 LQCGADSLGCDRLGCFNL 271
> YGL194c
Length=452
Score = 179 bits (455), Expect = 1e-45, Method: Composition-based stats.
Identities = 79/138 (57%), Positives = 105/138 (76%), Gaps = 1/138 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK+ SGFCYVND VL+ L LR+ R+LY+D+D+HHGDGV+EAFYT+ RV SFH
Sbjct 158 HHAKKNSPSGFCYVNDIVLSILNLLRYHPRILYIDIDLHHGDGVQEAFYTTDRVFTLSFH 217
Query 61 KY-GHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
KY G FFPGTG L +IG ++G +++NVPL +GIDD Y +LF+ ++D ++ ++P +V
Sbjct 218 KYNGEFFPGTGDLTEIGCDKGKHFALNVPLEDGIDDDSYINLFKSIVDPLIMTFKPTLIV 277
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+ DRLGCFNL
Sbjct 278 QQCGADSLGHDRLGCFNL 295
> At3g44680
Length=419
Score = 172 bits (436), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 83/138 (60%), Positives = 106/138 (76%), Gaps = 5/138 (3%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK +ASGFCY+ND VL LE L+ RVLY+D+D+HHGDGVEEAFY + RV+ SFH
Sbjct 133 HHAKKCDASGFCYINDLVLGILELLKHHPRVLYIDIDVHHGDGVEEAFYFTDRVMTVSFH 192
Query 61 KYG-HFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
K+G FFPGTG +++IG EG Y++NVPL +GIDD ++ LF+ V +E+Y+P A+V
Sbjct 193 KFGDKFFPGTGDVKEIGEREGKFYAINVPLKDGIDDSSFNRLFRTV----VEIYQPGAIV 248
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+A DRLGCFNL
Sbjct 249 LQCGADSLARDRLGCFNL 266
> SPAC3G9.07c
Length=434
Score = 172 bits (435), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 80/138 (57%), Positives = 104/138 (75%), Gaps = 1/138 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH K+ EASGFCYVND VLA L LRF RVLY+D+DIHHGDGV++AFY S RVL SFH
Sbjct 157 HHAKRGEASGFCYVNDIVLAILNMLRFFPRVLYIDIDIHHGDGVQQAFYESDRVLTVSFH 216
Query 61 KY-GHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
KY G FFP TG ++ G++ G +++NVPL +GI D+ Y+SLF+ +++ + ++P A+V
Sbjct 217 KYNGDFFPATGNFDENGVKGGKYFALNVPLEDGIGDEQYTSLFKSIIEPTINTFQPSAIV 276
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+ DRLG FNL
Sbjct 277 LQCGADSLGYDRLGVFNL 294
> CE01472
Length=507
Score = 164 bits (416), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 75/137 (54%), Positives = 99/137 (72%), Gaps = 0/137 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCYVND VL LE L++ RVLY+D+DIHHGDGV+EAF S RV+ SFH
Sbjct 161 HHAKKSEASGFCYVNDIVLGILELLKYHKRVLYIDIDIHHGDGVQEAFNNSDRVMTVSFH 220
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
++G +FPG+G++ D G+ G +++NVPL I D+ Y LF+ V+ + E + PEA+V
Sbjct 221 RFGQYFPGSGSIMDKGVGPGKYFAINVPLMAAIRDEPYLKLFESVISGVEENFNPEAIVL 280
Query 121 QCGADSVAGDRLGCFNL 137
QCG+DS+ DRLG F L
Sbjct 281 QCGSDSLCEDRLGQFAL 297
> Hs8923769
Length=377
Score = 151 bits (381), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 70/138 (50%), Positives = 97/138 (70%), Gaps = 1/138 (0%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK EASGFCY+ND VL L R R+LYVD+D+HHGDGVE+AF + +V+ S H
Sbjct 142 HHAKKDEASGFCYLNDAVLGILRLRRKFERILYVDLDLHHGDGVEDAFSFTSKVMTVSLH 201
Query 61 KYGH-FFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
K+ FFPGTG + D+GL +G YSVNVP+ +GI D+ Y + + V+ ++ + + P+AVV
Sbjct 202 KFSPGFFPGTGDVSDVGLGKGRYYSVNVPIQDGIQDEKYYQICESVLKEVYQAFNPKAVV 261
Query 120 FQCGADSVAGDRLGCFNL 137
Q GAD++AGD + FN+
Sbjct 262 LQLGADTIAGDPMCSFNM 279
> ECU09g0670
Length=344
Score = 144 bits (364), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 74/138 (53%), Positives = 91/138 (65%), Gaps = 6/138 (4%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH K SGFC+VND VLA LE L+ RV+Y+D+D+HHGDGVEEAF RVL S H
Sbjct 113 HHAHKAIPSGFCHVNDIVLAILELLKTYRRVMYIDIDVHHGDGVEEAFLECDRVLTLSLH 172
Query 61 KYGH-FFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVV 119
KYG FFP TG L G +VNVPL GIDD Y +F+ +++ + + P+A+V
Sbjct 173 KYGDGFFPETGTLITTGHR-----AVNVPLLSGIDDPSYRYIFEPIVESCVRRFSPDAIV 227
Query 120 FQCGADSVAGDRLGCFNL 137
QCGADS+ DRLGCFNL
Sbjct 228 LQCGADSLGEDRLGCFNL 245
> At5g35600
Length=409
Score = 137 bits (345), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 72/137 (52%), Positives = 89/137 (64%), Gaps = 4/137 (2%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFH 60
HH KK +ASGF YVND VLA LE L+ RVLY+++ HGD VEEAF + RV+ SFH
Sbjct 147 HHVKKDKASGFGYVNDVVLAILELLKSFKRVLYIEIGFPHGDEVEEAFKDTDRVMTVSFH 206
Query 61 KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAVVF 120
K G TG + D G +G YS+N PL +G+DD LF V+ + ME+Y PE +V
Sbjct 207 KVG----DTGDISDYGEGKGQYYSLNAPLKDGLDDFSLRGLFIPVIHRAMEIYEPEVIVL 262
Query 121 QCGADSVAGDRLGCFNL 137
QCGADS+AGD G FNL
Sbjct 263 QCGADSLAGDPFGTFNL 279
> YPR068c
Length=470
Score = 115 bits (287), Expect = 3e-26, Method: Composition-based stats.
Identities = 57/139 (41%), Positives = 84/139 (60%), Gaps = 5/139 (3%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK-HRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 59
HH K ASGFCY+ND VL + K +++ YVD D+HHGDGVE+AF S ++ S
Sbjct 210 HHAFKQRASGFCYINDVVLLIQRLRKAKLNKITYVDFDLHHGDGVEKAFQYSKQIQTISV 269
Query 60 HKYG-HFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAV 118
H Y FFPGTG+L D ++ + VN+PL G DD + ++++ ++E + PEA+
Sbjct 270 HLYEPGFFPGTGSLSDSRKDKNV---VNIPLKHGCDDNYLELIASKIVNPLIERHEPEAL 326
Query 119 VFQCGADSVAGDRLGCFNL 137
+ +CG D + GDR + L
Sbjct 327 IIECGGDGLLGDRFNEWQL 345
> SPBC800.03
Length=687
Score = 111 bits (278), Expect = 3e-25, Method: Composition-based stats.
Identities = 63/146 (43%), Positives = 82/146 (56%), Gaps = 9/146 (6%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFL-RFK---HRVLYVDVDIHHGDGVEEAFYTSPRVLC 56
HH + H+ GFC N+ + A L RF RVL VD DIHHG+G + AFY P VL
Sbjct 194 HHAEPHKPGGFCLFNNVSVTARSMLQRFPDKIKRVLIVDWDIHHGNGTQMAFYDDPNVLY 253
Query 57 CSFHKY--GHFFPGT--GALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIME 111
S H+Y G F+PGT G E+ G GLG +VN+P G+ D Y FQRV+ +
Sbjct 254 VSLHRYENGRFYPGTNYGCAENCGEGPGLGRTVNIPWSCAGMGDGDYIYAFQRVVMPVAY 313
Query 112 VYRPEAVVFQCGADSVAGDRLGCFNL 137
+ P+ V+ CG D+ AGD +G F L
Sbjct 314 EFDPDLVIVSCGFDAAAGDHIGQFLL 339
> Hs5174481
Length=1084
Score = 104 bits (260), Expect = 5e-23, Method: Composition-based stats.
Identities = 56/147 (38%), Positives = 86/147 (58%), Gaps = 10/147 (6%)
Query 1 HHGKKHEASGFCYVNDCVLAA--LEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH ++ GFCY N +AA L+ ++L VD D+HHG+G ++AFY+ P VL S
Sbjct 802 HHAEESTPMGFCYFNSVAVAAKLLQQRLSVSKILIVDWDVHHGNGTQQAFYSDPSVLYMS 861
Query 59 FHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGID----DQMYSSLFQRVMDQIMEV 112
H+Y G+FFPG+GA +++G G+G++VN+ G+D D Y + F+ V+ I
Sbjct 862 LHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLDPPMGDAEYLAAFRTVVMPIASE 921
Query 113 YRPEAVVFQCGADSVAG--DRLGCFNL 137
+ P+ V+ G D+V G LG +NL
Sbjct 922 FAPDVVLVSSGFDAVEGHPTPLGGYNL 948
> Hs17158039
Length=1011
Score = 100 bits (249), Expect = 7e-22, Method: Composition-based stats.
Identities = 48/138 (34%), Positives = 82/138 (59%), Gaps = 10/138 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++ A GFC+ N + A ++LR + ++L VD+D+HHG+G ++AFY P +L
Sbjct 782 HHAEESTAMGFCFFNSVAITA-KYLRDQLNISKILIVDLDVHHGNGTQQAFYADPSILYI 840
Query 58 SFHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQM----YSSLFQRVMDQIME 111
S H+Y G+FFPG+GA ++G G GY++N+ G+D M Y F+ ++ + +
Sbjct 841 SLHRYDEGNFFPGSGAPNEVGTGLGEGYNINIAWTGGLDPPMGDVEYLEAFRTIVKPVAK 900
Query 112 VYRPEAVVFQCGADSVAG 129
+ P+ V+ G D++ G
Sbjct 901 EFDPDMVLVSAGFDALEG 918
> 7292853
Length=1235
Score = 97.8 bits (242), Expect = 6e-21, Method: Composition-based stats.
Identities = 52/139 (37%), Positives = 80/139 (57%), Gaps = 11/139 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK----HRVLYVDVDIHHGDGVEEAFYTSPRVLC 56
HH + + A GFC+ N +AA + LR + R+L VD D+HHG+G ++AFY SP +L
Sbjct 967 HHAEANLAMGFCFFNSIAIAA-KLLRQRMPEVRRILIVDWDVHHGNGTQQAFYQSPDILY 1025
Query 57 CSFHKY--GHFFPGTGALEDIGLEEGLGYSVNV----PLHEGIDDQMYSSLFQRVMDQIM 110
S H++ G+FFPGTG + G GLG++VN+ L+ + D Y + F+ V+ I
Sbjct 1026 LSIHRHDDGNFFPGTGGPTECGSGAGLGFNVNISWSGALNPPLGDAEYIAAFRTVVMPIA 1085
Query 111 EVYRPEAVVFQCGADSVAG 129
+ P+ V+ G D+ G
Sbjct 1086 RSFNPDIVLVSSGFDAATG 1104
> Hs13259524
Length=912
Score = 95.1 bits (235), Expect = 4e-20, Method: Composition-based stats.
Identities = 49/137 (35%), Positives = 76/137 (55%), Gaps = 8/137 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAA--LEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH A GFC+ N +A L+ ++L VD D+HHG+G ++ FY P VL S
Sbjct 629 HHADHSTAMGFCFFNSVAIACRQLQQQSKASKILIVDWDVHHGNGTQQTFYQDPSVLYIS 688
Query 59 FHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQM----YSSLFQRVMDQIMEV 112
H++ G+FFPG+GA++++G G G++VNV G+D M Y + F+ V+ I
Sbjct 689 LHRHDDGNFFPGSGAVDEVGAGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIARE 748
Query 113 YRPEAVVFQCGADSVAG 129
+ P+ V+ G D+ G
Sbjct 749 FSPDLVLVSAGFDAAEG 765
> Hs13259522
Length=952
Score = 95.1 bits (235), Expect = 4e-20, Method: Composition-based stats.
Identities = 49/137 (35%), Positives = 76/137 (55%), Gaps = 8/137 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAA--LEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH A GFC+ N +A L+ ++L VD D+HHG+G ++ FY P VL S
Sbjct 669 HHADHSTAMGFCFFNSVAIACRQLQQQSKASKILIVDWDVHHGNGTQQTFYQDPSVLYIS 728
Query 59 FHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQM----YSSLFQRVMDQIMEV 112
H++ G+FFPG+GA++++G G G++VNV G+D M Y + F+ V+ I
Sbjct 729 LHRHDDGNFFPGSGAVDEVGAGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIARE 788
Query 113 YRPEAVVFQCGADSVAG 129
+ P+ V+ G D+ G
Sbjct 789 FSPDLVLVSAGFDAAEG 805
> Hs4885531
Length=1122
Score = 93.2 bits (230), Expect = 1e-19, Method: Composition-based stats.
Identities = 57/138 (41%), Positives = 82/138 (59%), Gaps = 10/138 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++ A GFC+ N + A + L+ K +VL VD DIHHG+G ++AFY P VL
Sbjct 832 HHAEESTAMGFCFFNSVAITA-KLLQQKLNVGKVLIVDWDIHHGNGTQQAFYNDPSVLYI 890
Query 58 SFHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGID----DQMYSSLFQRVMDQIME 111
S H+Y G+FFPG+GA E++G G+GY+VNV G+D D Y + F+ V+ I
Sbjct 891 SLHRYDNGNFFPGSGAPEEVGGGPGVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAH 950
Query 112 VYRPEAVVFQCGADSVAG 129
+ P+ V+ G D+V G
Sbjct 951 EFSPDVVLVSAGFDAVEG 968
> Hs21237799
Length=1037
Score = 93.2 bits (230), Expect = 1e-19, Method: Composition-based stats.
Identities = 57/138 (41%), Positives = 82/138 (59%), Gaps = 10/138 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++ A GFC+ N + A + L+ K +VL VD DIHHG+G ++AFY P VL
Sbjct 747 HHAEESTAMGFCFFNSVAITA-KLLQQKLNVGKVLIVDWDIHHGNGTQQAFYNDPSVLYI 805
Query 58 SFHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHEGID----DQMYSSLFQRVMDQIME 111
S H+Y G+FFPG+GA E++G G+GY+VNV G+D D Y + F+ V+ I
Sbjct 806 SLHRYDNGNFFPGSGAPEEVGGGPGVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAH 865
Query 112 VYRPEAVVFQCGADSVAG 129
+ P+ V+ G D+V G
Sbjct 866 EFSPDVVLVSAGFDAVEG 883
> At1g08460
Length=377
Score = 92.0 bits (227), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 50/143 (34%), Positives = 78/143 (54%), Gaps = 6/143 (4%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKH--RVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH + +A G+C++N+ LA L RV +D+D+H+G+G E FYTS +VL S
Sbjct 144 HHSQPTQADGYCFLNNAALAVKLALNSGSCSRVAVIDIDVHYGNGTAEGFYTSDKVLTVS 203
Query 59 FH----KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYR 114
H +G P G+++++G + GLGY++NVPL G D+ Y ++ + +
Sbjct 204 LHMNHGSWGSSHPQKGSIDELGEDVGLGYNLNVPLPNGTGDRGYEYAMNELVVPAVRRFG 263
Query 115 PEAVVFQCGADSVAGDRLGCFNL 137
P+ VV G DS A D G +L
Sbjct 264 PDMVVLVVGQDSSAFDPNGRQSL 286
> At3g18520
Length=561
Score = 91.3 bits (225), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 55/138 (39%), Positives = 76/138 (55%), Gaps = 4/138 (2%)
Query 1 HHGKKHEASGFC-YVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 59
HH A GFC + N V A + +VL VD D+HHG+G +E F + VL S
Sbjct 285 HHAGVRHAMGFCLHNNAAVAALVAQAAGAKKVLIVDWDVHHGNGTQEIFEQNKSVLYISL 344
Query 60 HKY--GHFFPGTGALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIMEVYRPE 116
H++ G+F+PGTGA +++G G GY VNVP G+ D+ Y FQ V+ I + P+
Sbjct 345 HRHEGGNFYPGTGAADEVGSNGGEGYCVNVPWSCGGVGDKDYIFAFQHVVLPIASAFSPD 404
Query 117 AVVFQCGADSVAGDRLGC 134
V+ G D+ GD LGC
Sbjct 405 FVIISAGFDAARGDPLGC 422
> CE08062
Length=796
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 44/137 (32%), Positives = 81/137 (59%), Gaps = 9/137 (6%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKH-----RVLYVDVDIHHGDGVEEAFYTSPRVL 55
HH + +A GFC+ N+ V A++ L+ K+ ++ +D D+HHG+G + +F P VL
Sbjct 537 HHAEHEQAMGFCFFNN-VAVAVKVLQTKYPAQCAKIAIIDWDVHHGNGTQLSFENDPNVL 595
Query 56 CCSFHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIMEV 112
S H++ G+FFPGTG++ ++G + G +VNVP + + D Y + ++ V++ +M
Sbjct 596 YMSLHRHDKGNFFPGTGSVTEVGKNDAKGLTVNVPFSGDVMRDPEYLAAWRTVIEPVMAS 655
Query 113 YRPEAVVFQCGADSVAG 129
+ P+ ++ G D+ G
Sbjct 656 FCPDFIIVSAGFDACHG 672
> 7293057
Length=883
Score = 87.8 bits (216), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 51/140 (36%), Positives = 79/140 (56%), Gaps = 7/140 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLR--FKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH ++ GFC N+ +AA +R RVL VD D+HHG+G + F ++P+VL S
Sbjct 588 HHAEQDHPHGFCIFNNVAIAAQYAIRDFGLERVLIVDWDVHHGNGTQHIFESNPKVLYIS 647
Query 59 FHKYGH--FFPG--TGALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIMEVY 113
H+Y H FFP G + +G G G++VN+P + +G+ D Y+ FQ+++ I +
Sbjct 648 LHRYEHGSFFPKGPDGNFDVVGKGAGRGFNVNIPWNKKGMGDLEYALAFQQLIMPIAYEF 707
Query 114 RPEAVVFQCGADSVAGDRLG 133
P+ V+ G D+ GD LG
Sbjct 708 NPQLVLVSAGFDAAIGDPLG 727
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 47/137 (34%), Positives = 77/137 (56%), Gaps = 9/137 (6%)
Query 1 HHGKKHEASGFCYVNDCVLA---ALEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH K E +G+C+ N+ LA AL+ + + R+L +D D+HHG G + FY PRV+
Sbjct 161 HHAMKAEYNGYCFFNNVALATQHALDVHKLQ-RILIIDYDVHHGQGTQRFFYNDPRVVYF 219
Query 58 SFHKYGH--FFP--GTGALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIMEV 112
S H++ H F+P IG G GY+ NVPL+ G+ + Y ++FQ+++ +
Sbjct 220 SIHRFEHGSFWPHLHESDYHAIGSGAGTGYNFNVPLNATGMTNGDYLAIFQQLLLPVALE 279
Query 113 YRPEAVVFQCGADSVAG 129
++PE ++ G D+ G
Sbjct 280 FQPELIIVSAGYDAALG 296
> CE20772
Length=863
Score = 85.9 bits (211), Expect = 2e-17, Method: Composition-based stats.
Identities = 52/142 (36%), Positives = 76/142 (53%), Gaps = 11/142 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKH---RVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++SGFC N+ +AA ++ + +H RVL +D D+HHG+G +E FY V+
Sbjct 560 HHASASKSSGFCIFNNVAVAA-KYAQRRHKAKRVLILDWDVHHGNGTQEIFYEDSNVMYM 618
Query 58 SFHKY--GHFFP--GTGALEDIGLEEGLGYSVNVPLHEGID--DQMYSSLFQRVMDQIME 111
S H++ G+F+P D+G G G SVNVP G+ D Y FQRV+ I
Sbjct 619 SIHRHDKGNFYPIGEPKDYSDVGEGAGEGMSVNVPF-SGVQMGDNEYQMAFQRVIMPIAY 677
Query 112 VYRPEAVVFQCGADSVAGDRLG 133
+ P+ V+ G D+ D LG
Sbjct 678 QFNPDLVLISAGFDAAVDDPLG 699
Score = 85.1 bits (209), Expect = 3e-17, Method: Composition-based stats.
Identities = 51/139 (36%), Positives = 71/139 (51%), Gaps = 6/139 (4%)
Query 1 HHGKKHEASGFCYVNDCVLAALE-FLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 59
HH GFC N+ AA E F R+L VD+D+HHG G + FY RVL S
Sbjct 145 HHADSVSPCGFCLFNNVAQAAEEAFFSGAERILIVDLDVHHGHGTQRIFYDDKRVLYFSI 204
Query 60 HKYGH--FFPG--TGALEDIGLEEGLGYSVNVPLHE-GIDDQMYSSLFQRVMDQIMEVYR 114
H++ H F+P + IG +GLGY+ N+ L+E G D Y S+ V+ + +
Sbjct 205 HRHEHGLFWPHLPESDFDHIGSGKGLGYNANLALNETGCTDSDYLSIIFHVLLPLATQFD 264
Query 115 PEAVVFQCGADSVAGDRLG 133
P V+ G D++ GD LG
Sbjct 265 PHFVIISAGFDALLGDPLG 283
> Hs20070354
Length=669
Score = 85.9 bits (211), Expect = 2e-17, Method: Composition-based stats.
Identities = 49/140 (35%), Positives = 78/140 (55%), Gaps = 7/140 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK--HRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HHG++ A+GFC N+ +AA + HR+L VD D+HHG G++ F P VL S
Sbjct 134 HHGQRAAANGFCVFNNVAIAAAHAKQKHGLHRILVVDWDVHHGQGIQYLFEDDPSVLYFS 193
Query 59 FHKYGH--FFP--GTGALEDIGLEEGLGYSVNVPLHE-GIDDQMYSSLFQRVMDQIMEVY 113
+H+Y H F+P + +G +GLG++VN+P ++ G+ + Y + F ++ + +
Sbjct 194 WHRYEHGRFWPFLRESDADAVGRGQGLGFTVNLPWNQVGMGNADYVAAFLHLLLPLAFEF 253
Query 114 RPEAVVFQCGADSVAGDRLG 133
PE V+ G DS GD G
Sbjct 254 DPELVLVSAGFDSAIGDPEG 273
> CE25887
Length=955
Score = 85.1 bits (209), Expect = 3e-17, Method: Composition-based stats.
Identities = 52/142 (36%), Positives = 76/142 (53%), Gaps = 11/142 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKH---RVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++SGFC N+ +AA ++ + +H RVL +D D+HHG+G +E FY V+
Sbjct 560 HHASASKSSGFCIFNNVAVAA-KYAQRRHKAKRVLILDWDVHHGNGTQEIFYEDSNVMYM 618
Query 58 SFHKY--GHFFP--GTGALEDIGLEEGLGYSVNVPLHEGID--DQMYSSLFQRVMDQIME 111
S H++ G+F+P D+G G G SVNVP G+ D Y FQRV+ I
Sbjct 619 SIHRHDKGNFYPIGEPKDYSDVGEGAGEGMSVNVPF-SGVQMGDNEYQMAFQRVIMPIAY 677
Query 112 VYRPEAVVFQCGADSVAGDRLG 133
+ P+ V+ G D+ D LG
Sbjct 678 QFNPDLVLISAGFDAAVDDPLG 699
Score = 84.3 bits (207), Expect = 6e-17, Method: Composition-based stats.
Identities = 51/139 (36%), Positives = 71/139 (51%), Gaps = 6/139 (4%)
Query 1 HHGKKHEASGFCYVNDCVLAALE-FLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSF 59
HH GFC N+ AA E F R+L VD+D+HHG G + FY RVL S
Sbjct 145 HHADSVSPCGFCLFNNVAQAAEEAFFSGAERILIVDLDVHHGHGTQRIFYDDKRVLYFSI 204
Query 60 HKYGH--FFPG--TGALEDIGLEEGLGYSVNVPLHE-GIDDQMYSSLFQRVMDQIMEVYR 114
H++ H F+P + IG +GLGY+ N+ L+E G D Y S+ V+ + +
Sbjct 205 HRHEHGLFWPHLPESDFDHIGSGKGLGYNANLALNETGCTDSDYLSIIFHVLLPLATQFD 264
Query 115 PEAVVFQCGADSVAGDRLG 133
P V+ G D++ GD LG
Sbjct 265 PHFVIISAGFDALLGDPLG 283
> Hs13128864
Length=1215
Score = 84.7 bits (208), Expect = 4e-17, Method: Composition-based stats.
Identities = 51/141 (36%), Positives = 71/141 (50%), Gaps = 8/141 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKH---RVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++ A GFC+ N +AA R+L VD D+HHG+G + F P VL
Sbjct 610 HHAEQDAACGFCFFNSVAVAARHAQTISGHALRILIVDWDVHHGNGTQHMFEDDPSVLYV 669
Query 58 SFHKYGH--FFP--GTGALEDIGLEEGLGYSVNVPLH-EGIDDQMYSSLFQRVMDQIMEV 112
S H+Y H FFP GA IG G G++VNV + + D Y + + R++ I
Sbjct 670 SLHRYDHGTFFPMGDEGASSQIGRAAGTGFTVNVAWNGPRMGDADYLAAWHRLVLPIAYE 729
Query 113 YRPEAVVFQCGADSVAGDRLG 133
+ PE V+ G D+ GD LG
Sbjct 730 FNPELVLVSAGFDAARGDPLG 750
Score = 79.3 bits (194), Expect = 2e-15, Method: Composition-based stats.
Identities = 49/141 (34%), Positives = 73/141 (51%), Gaps = 9/141 (6%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHR---VLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH + G+C N +AA + + KHR VL VD D+HHG G + F P VL
Sbjct 215 HHAQHSLMDGYCMFNHVAVAA-RYAQQKHRIRRVLIVDWDVHHGQGTQFTFDQDPSVLYF 273
Query 58 SFHKY--GHFFPGTGA--LEDIGLEEGLGYSVNVPLHE-GIDDQMYSSLFQRVMDQIMEV 112
S H+Y G F+P A G +G GY++NVP ++ G+ D Y + F V+ +
Sbjct 274 SIHRYEQGRFWPHLKASNWSTTGFGQGQGYTINVPWNQVGMRDADYIAAFLHVLLPVALE 333
Query 113 YRPEAVVFQCGADSVAGDRLG 133
++P+ V+ G D++ GD G
Sbjct 334 FQPQLVLVAAGFDALQGDPKG 354
> YNL021w
Length=706
Score = 83.2 bits (204), Expect = 1e-16, Method: Composition-based stats.
Identities = 45/143 (31%), Positives = 79/143 (55%), Gaps = 11/143 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLR----FKHRVLYVDVDIHHGDGVEEAFYTSPRVLC 56
HH + A GFC ++ +AA L+ R++ +D DIHHG+G +++FY +VL
Sbjct 205 HHAEPQAAGGFCLFSNVAVAAKNILKNYPESVRRIMILDWDIHHGNGTQKSFYQDDQVLY 264
Query 57 CSFHKY--GHFFPGT--GALEDIGLEEGLGYSVNV--PLHEGIDDQMYSSLFQRVMDQIM 110
S H++ G ++PGT G + G +G G++ N+ P+ G+ D Y F++V+ +
Sbjct 265 VSLHRFEMGKYYPGTIQGQYDQTGEGKGEGFNCNITWPVG-GVGDAEYMWAFEQVVMPMG 323
Query 111 EVYRPEAVVFQCGADSVAGDRLG 133
++P+ V+ G D+ GD +G
Sbjct 324 REFKPDLVIISSGFDAADGDTIG 346
> At4g33470
Length=359
Score = 77.4 bits (189), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 68/135 (50%), Gaps = 3/135 (2%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK--HRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH GFC + +AA R R+ +D D+HHG+G +AF P + S
Sbjct 137 HHAVPKGPMGFCVFGNVAIAARHAQRTHGLKRIFIIDFDVHHGNGTNDAFTEDPDIFFLS 196
Query 59 FHKYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPEAV 118
H+ G + PGTG + DIG +G G ++N+PL G D ++F+ ++ + ++P+ +
Sbjct 197 THQDGSY-PGTGKISDIGKGKGEGTTLNLPLPGGSGDIAMRTVFEEIIVPCAQRFKPDII 255
Query 119 VFQCGADSVAGDRLG 133
+ G D+ D L
Sbjct 256 LVSAGYDAHVLDPLA 270
> Hs17158041
Length=879
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 33/80 (41%), Positives = 53/80 (66%), Gaps = 6/80 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK---HRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH ++ A GFC+ N + A ++LR + ++L VD+D+HHG+G ++AFY P +L
Sbjct 782 HHAEESTAMGFCFFNSVAITA-KYLRDQLNISKILIVDLDVHHGNGTQQAFYADPSILYI 840
Query 58 SFHKY--GHFFPGTGALEDI 75
S H+Y G+FFPG+GA ++
Sbjct 841 SLHRYDEGNFFPGSGAPNEV 860
> CE24411
Length=517
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 41/138 (29%), Positives = 68/138 (49%), Gaps = 8/138 (5%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYVDVDIHHGDGVEEAF--YTSPRVLCCS 58
HH E GFC N+ +AA ++ +VL VD D+H G+G +E V S
Sbjct 137 HHAMPDEGCGFCIFNNVAIAAKAAIQNGQKVLIVDYDVHAGNGTQECVEQMGEGNVQLIS 196
Query 59 FHKY--GHFFPGTGALEDIGLEEGLGYSVNVPLHE-GIDDQMYSSLFQRVMDQIMEVYRP 115
H+Y GHF+P + G+ ++N+PL+ G+ D Y +LF ++ + ++P
Sbjct 197 IHRYENGHFWPN---MPQTGIYHNYKNTINLPLNTIGLTDADYHALFTHIILPTIHAFQP 253
Query 116 EAVVFQCGADSVAGDRLG 133
+ ++ G D+ GD G
Sbjct 254 DLLLVSSGFDASIGDPEG 271
> At5g61060
Length=717
Score = 66.6 bits (161), Expect = 1e-11, Method: Composition-based stats.
Identities = 42/154 (27%), Positives = 70/154 (45%), Gaps = 28/154 (18%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK-----HRVLYVDVDIHHGDGVEEAFYTSPR-- 53
HH + EA GFC N+ +AA L + ++L VD D+HHG+G ++ F+ PR
Sbjct 157 HHAEADEAMGFCLFNNVAVAASFLLNERPDLGVKKILIVDWDVHHGNGTQKMFWKDPREP 216
Query 54 ---------------VLCCSF---HKYGHFFPG--TGALEDIGLEEGLGYSVNVPLHEG- 92
V +F H+YG F+P G +G G G+++NVP +G
Sbjct 217 TFYSITPLLDFYAWKVTGNAFLVRHEYGGFYPAGDDGDYNMVGEGPGEGFNINVPWDQGR 276
Query 93 IDDQMYSSLFQRVMDQIMEVYRPEAVVFQCGADS 126
D Y + + ++ + + P+ + G D+
Sbjct 277 CGDADYLAAWDHILIPVAREFNPDVIFLSAGFDA 310
> At5g61070
Length=714
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 45/182 (24%), Positives = 71/182 (39%), Gaps = 57/182 (31%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLR----------------------FKH--------- 29
HH + EA GFC N+ +AA L F H
Sbjct 190 HHAESDEAMGFCLFNNVAVAASFLLNERVMTSFDFCDAFSISYLLSNFFGHIPLVRTFVL 249
Query 30 ---------------RVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYGHFFP--GTGAL 72
++L VD DIHHG+ AF T H +G F+P G
Sbjct 250 MLCGINRMQPDLDVKKILIVDWDIHHGNVTGNAFVTR--------HDHGSFYPFGDDGDF 301
Query 73 EDIGLEEGLGYSVNVPLHEG-IDDQMYSSLFQRVMDQIMEVYRPEAVVFQCGADSVAGDR 131
+G G G+++NVP +G D Y +++ ++ + + ++P+ ++ G D+ GD
Sbjct 302 NMVGEGPGEGFNINVPWEQGGCGDADYLAVWNHILIPVTKEFKPDIILLSAGFDAAIGDP 361
Query 132 LG 133
LG
Sbjct 362 LG 363
> CE20787
Length=782
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/132 (29%), Positives = 65/132 (49%), Gaps = 10/132 (7%)
Query 1 HHGKKHEASGFCYVNDCVLAA---LEFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCC 57
HH A G+C N+ +AA +E L K +V VD D H G+G ++ R+
Sbjct 407 HHSYGKVAQGYCIFNNVAIAAKYAIEKLEVK-KVAIVDFDYHAGNGTYQSVKDESRIHLT 465
Query 58 SFH--KYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQM-YSSLFQRVMDQIMEVYR 114
SFH +G F+P + D ++ VPL+ ++ + Y S+F V+ ++E ++
Sbjct 466 SFHGHHFGAFWPFS---RDYDYATNSQNTLFVPLNGTMNSEGDYVSVFHHVLLPMLEQFQ 522
Query 115 PEAVVFQCGADS 126
PE ++ G DS
Sbjct 523 PELILISAGFDS 534
> Hs13376228
Length=347
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 63/141 (44%), Gaps = 13/141 (9%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK----HRVLYVDVDIHHGDGVEEAFYTSPRVLC 56
HH GFC D LA ++FL + R +D+D H G+G E F RV
Sbjct 142 HHCSSDRGGGFCAYADITLA-IKFLFERVEGISRATIIDLDAHQGNGHERDFMDDKRVYI 200
Query 57 CSFHKYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRPE 116
+ H +PG D ++ + V L G +D Y +R + + ++ + P+
Sbjct 201 MDVYNR-HIYPG-----DRFAKQAIRRKVE--LEWGTEDDEYLDKVERNIKKSLQEHLPD 252
Query 117 AVVFQCGADSVAGDRLGCFNL 137
VV+ G D + GDRLG ++
Sbjct 253 VVVYNAGTDILEGDRLGGLSI 273
> 7301219
Length=343
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 39/138 (28%), Positives = 58/138 (42%), Gaps = 14/138 (10%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFK----HRVLYVDVDIHHGDGVEEAFYTSPRVLC 56
HH + GFC D L + + R++ VD+D H G+G E F V
Sbjct 158 HHCCSYRGGGFCPYADISLLIVRLFEQEPFRVRRIMIVDLDAHQGNGHERDFNNVAAVYI 217
Query 57 CSFHKYGHF-FPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEVYRP 115
F Y F +P D +E + +V L +D Y +R + Q + +RP
Sbjct 218 --FDMYNAFVYP-----RDHVAKESIRCAVE--LRNYTEDGFYLRQLKRCLMQSLAEFRP 268
Query 116 EAVVFQCGADSVAGDRLG 133
+ VV+ G D + GD LG
Sbjct 269 DMVVYNAGTDVLEGDPLG 286
> At5g26040
Length=316
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 58/143 (40%), Gaps = 19/143 (13%)
Query 1 HHGKKHEASGFCYVNDCVLAA-LEFLRFK-HRVLYVDVDIHHGDGVEEAFYTSPRVLCCS 58
HH GFC D L FLR + RV+ +D+D H G+G E +L
Sbjct 130 HHCTAERGGGFCAFADISLCIHFAFLRLRISRVMIIDLDAHQGNGHETDL--GDDILSML 187
Query 59 FHKYGHFFPGTGALEDIGLEEGLGYSVNVPLHEGIDDQMYSSLFQRVMDQIMEV----YR 114
+ F L+D + V V D+ + R +D+ +EV ++
Sbjct 188 AFEVRCIF-----LQDYRARRFIDQKVEVMSGTTTDE------YLRKLDEALEVASRNFQ 236
Query 115 PEAVVFQCGADSVAGDRLGCFNL 137
PE V++ G D + GD LG +
Sbjct 237 PELVIYNAGTDILDGDPLGLLKI 259
> YPL116w
Length=697
Score = 35.4 bits (80), Expect = 0.033, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 1 HHGKKHEASGFCYVNDCVLAALEFLRFKHRVLYV---DVDIHHGDGVEE 46
HH SGFC +N+ +A +E+ + V +V D D+HHGDG ++
Sbjct 195 HHCHYGTPSGFCLLNNAHVA-IEYAYDTYNVTHVVVLDFDLHHGDGTQD 242
> 7290780
Length=362
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query 23 EFLRFKHRVLYVDVDIHHGDGVEEAFYTSPRVLCCSFHKYG 63
EFL + ++ + D+ V+ Y PRV C+FHKYG
Sbjct 205 EFLSYGTNIMKLS-DVPQEQRVDPMVYVFPRVTKCTFHKYG 244
> 7295572
Length=372
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 3/42 (7%)
Query 23 EFLRFKHRVL-YVDVDIHHGDGVEEAFYTSPRVLCCSFHKYG 63
EF+ F +V+ Y++ D D ++ Y PR+ C+F KYG
Sbjct 206 EFITFGLKVIDYMETD--QEDRMDPMIYIFPRMTKCTFFKYG 245
Lambda K H
0.326 0.144 0.462
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40