bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
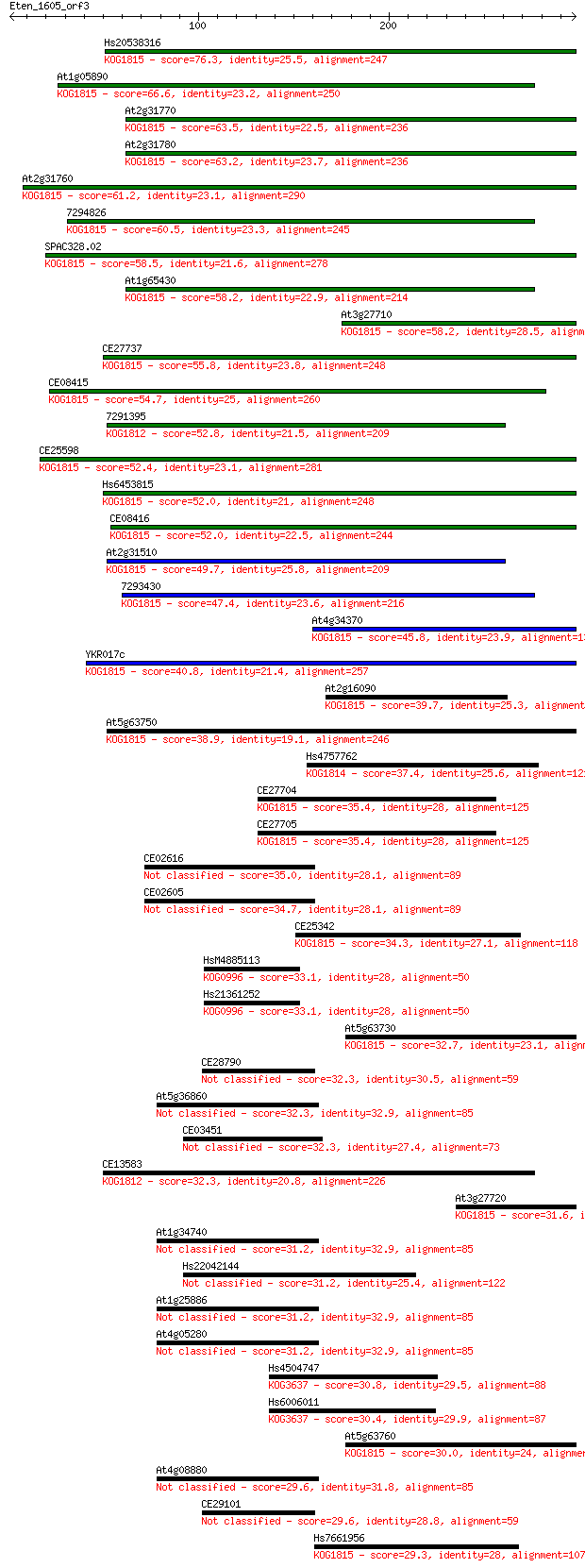
Query= Eten_1605_orf3
Length=297
Score E
Sequences producing significant alignments: (Bits) Value
Hs20538316 76.3 8e-14
At1g05890 66.6 7e-11
At2g31770 63.5 5e-10
At2g31780 63.2 6e-10
At2g31760 61.2 3e-09
7294826 60.5 4e-09
SPAC328.02 58.5 2e-08
At1g65430 58.2 2e-08
At3g27710 58.2 2e-08
CE27737 55.8 9e-08
CE08415 54.7 2e-07
7291395 52.8 8e-07
CE25598 52.4 1e-06
Hs6453815 52.0 1e-06
CE08416 52.0 2e-06
At2g31510 49.7 7e-06
7293430 47.4 4e-05
At4g34370 45.8 1e-04
YKR017c 40.8 0.004
At2g16090 39.7 0.008
At5g63750 38.9 0.013
Hs4757762 37.4 0.041
CE27704 35.4 0.16
CE27705 35.4 0.16
CE02616 35.0 0.19
CE02605 34.7 0.22
CE25342 34.3 0.33
HsM4885113 33.1 0.71
Hs21361252 33.1 0.71
At5g63730 32.7 0.88
CE28790 32.3 1.2
At5g36860 32.3 1.3
CE03451 32.3 1.3
CE13583 32.3 1.4
At3g27720 31.6 2.3
At1g34740 31.2 2.5
Hs22042144 31.2 2.7
At1g25886 31.2 2.7
At4g05280 31.2 3.1
Hs4504747 30.8 3.6
Hs6006011 30.4 4.1
At5g63760 30.0 6.0
At4g08880 29.6 7.4
CE29101 29.6 9.1
Hs7661956 29.3 9.2
> Hs20538316
Length=1089
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 63/252 (25%), Positives = 103/252 (40%), Gaps = 21/252 (8%)
Query 51 PESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRL--- 107
P+ L R D LI ETA ++ LLR DW + L++ W ++ ++ +
Sbjct 230 PQDLRRLKDMLIVETADMLQAPLFTAEALLRAHDWDREKLLEAWMSNPENCCQRSGVQMP 289
Query 108 -PPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACV 166
PP + P +P P + S++ +P ++S D C + C
Sbjct 290 TPPPSGYNAWDTLP-------SPRTPRTTRSSVTSP---DEISLSPGDLDTSLCDICMCS 339
Query 167 VPLTET-FALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRG 225
+ + E + C H + W +L+ + EG A + CP C +LV +D+
Sbjct 340 ISVFEDPVDMPCGHDFCRGCWESFLNLKIQEG--EAHNIFCPAYDCFQLVPVDIIESVVS 397
Query 226 PSAVARLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALV 285
R F + FV + A+ WCP+P C +AV L G + S SD+ + +
Sbjct 398 KEMDKRYLQFDIKAFVENNPAIKWCPTPGCDRAVRLTKQGSNTSGSDTL----SFPLLRA 453
Query 286 ADVTCSCGARFC 297
V C G FC
Sbjct 454 PAVDCGKGHLFC 465
> At1g05890
Length=545
Score = 66.6 bits (161), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 58/261 (22%), Positives = 94/261 (36%), Gaps = 51/261 (19%)
Query 26 YDDDELMES----AVASPQSKGLIPRYHSPE-SLDRRIDELIRETAALMGLNEAFVGILL 80
YDD++ E+ A +I Y S E + R ++ + +A++ + + LL
Sbjct 23 YDDNDSDETDFGFGEADTDDAAIIASYRSKEEDIRRHQNDDVGRVSAVLSITDVEASTLL 82
Query 81 RKFDWRSDALIQEWFNDSAAVLRK-GRLP-PVVREQDREDAPGTAECESAPAEPEASDSA 138
+ W + EWF D V R G L PVV D
Sbjct 83 LHYHWSVSKVNDEWFADEERVRRTVGILEGPVVTTPD----------------------- 119
Query 139 LKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTEGP 198
G F C + L E ++ C H + W GY+ + +GP
Sbjct 120 ----------------GREFTCGICFDSYTLEEIVSVSCGHPFCATCWTGYISTTINDGP 163
Query 199 QRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSPMCQQA 258
L +CP P C + D+ ++ + + +V + + WCP+P C+ A
Sbjct 164 G-CLMLKCPDPSCPAAIGRDMIDKLASKEDKEKYYRYFLRSYVEVNREMKWCPAPGCEHA 222
Query 259 VELVPA----GVSCSDSDSFC 275
++ VSC S SFC
Sbjct 223 IDFAGGTESYDVSCLCSHSFC 243
> At2g31770
Length=543
Score = 63.5 bits (153), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 53/238 (22%), Positives = 82/238 (34%), Gaps = 51/238 (21%)
Query 62 IRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQDREDAPG 121
I ++++ L++ V +LL ++W + EWF D + + L
Sbjct 59 IERVSSILSLSQVEVIVLLLHYNWCVSKVEDEWFTDEERIRKAVGL-------------- 104
Query 122 TAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLT--ETFALKCSH 179
LK P + +K++ C T E + C H
Sbjct 105 -----------------LKEPVVDFNGGEKDKKCRKVNIQCGICFESYTREEIARVSCGH 147
Query 180 RYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQ 239
Y W GY+ + +GP L +CP P C V D+ + +
Sbjct 148 PYCKTCWAGYITTKIEDGPG-CLRVKCPEPSCSAAVGKDMIEDVTETKVNEKYSRYILRS 206
Query 240 FVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCGARFC 297
+V + WCPSP C AVE S+S S+ DV+C C RFC
Sbjct 207 YVEDGKKIKWCPSPGCGYAVEF-----GGSESSSY------------DVSCLCSYRFC 247
> At2g31780
Length=542
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 56/238 (23%), Positives = 86/238 (36%), Gaps = 58/238 (24%)
Query 62 IRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRK-GRLP-PVVREQDREDA 119
I + + ++ +++ +LL + W L EWF D + + G L PVV D
Sbjct 77 IEQVSTVLSVSQVESIVLLLHYHWCVSKLEDEWFTDEERIRKTVGILKEPVV------DV 130
Query 120 PGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSH 179
GT VD+ +C + E + C H
Sbjct 131 NGT----------------------EVDI----------QCGICFESYTRKEIARVSCGH 158
Query 180 RYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQ 239
Y W GY+ + +GP L +CP P C +V D+ + + +
Sbjct 159 PYCKTCWTGYITTKIEDGPG-CLRVKCPEPSCYAVVGQDMIDEVTEKKDKDKYYRYFLRS 217
Query 240 FVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCGARFC 297
+V + WCPSP C+ AVE GV+ S S DV+C C +FC
Sbjct 218 YVEDGKKMKWCPSPGCEYAVEF---GVNGSSS--------------YDVSCLCSYKFC 258
> At2g31760
Length=514
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 67/292 (22%), Positives = 106/292 (36%), Gaps = 71/292 (24%)
Query 8 EVESDGACAEGCELDEFIYDDDELMESAVASPQSKGLIPRYHSPESLDRRIDELIRETAA 67
E SDG E Y+ + +++ + S +S +I E L + D++ R +
Sbjct 16 ENNSDGGGNES-------YNYNAAVDTIILSEKSYVIIKE---EEILKLQRDDIER-VST 64
Query 68 LMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRK-GRLP-PVVREQDREDAPGTAEC 125
++ L++ +LL + W L EWF D + + G L PVV D GT
Sbjct 65 ILFLSQVEAIVLLLHYHWCVSKLEDEWFTDEERIRKTVGILKEPVV------DVNGT--- 115
Query 126 ESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAA 185
VD+ +C + E ++ C H Y
Sbjct 116 -------------------EVDI----------QCGICFESYTRKEIASVSCGHPYCKTC 146
Query 186 WIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQFVGKHL 245
W GY+ + +GP L +CP P C +V D+ + + + +V
Sbjct 147 WTGYITTKIEDGPG-CLRVKCPEPSCYAVVGQDMIDEVTEKKDKDKYYRYFLRSYVEDGK 205
Query 246 AVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCGARFC 297
+ WCPSP C+ AVE +S + DV C C RFC
Sbjct 206 KMKWCPSPGCECAVEF-------GESSGY------------DVACLCSYRFC 238
> 7294826
Length=511
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 57/252 (22%), Positives = 92/252 (36%), Gaps = 41/252 (16%)
Query 31 LMESAVASPQSKGLIPRYHSPESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDAL 90
L+ SP+++ + + S + + + +I E ++ L I+L F W ++L
Sbjct 39 LLPENSNSPETEDFVYKVLSVDQIVQHQRNIIDEVNNVLNLPPQVTRIILNHFKWDKESL 98
Query 91 IQEWF--NDSAAVLRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDV 148
+ +F N R L P ++ +RE A T+ A+ C
Sbjct 99 FENYFESNPKDFFQRAHVLNPFEKKIERESAASTS-------------CAIPQLCG---- 141
Query 149 SKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYL-DALVTEGPQRALDARCP 207
C E L C H + A W YL + +EG + +CP
Sbjct 142 ---------------ICFCSCDELIGLGCGHNFCAACWKQYLANKTCSEGLANTI--KCP 184
Query 208 FPGCLELVTMDVWNRFRGPSAVA-RLRMFAREQFVGKHLAVAWCPSPMCQQAVELV---P 263
C LV + + S V R + FV ++ + WCP+P C AV+ V P
Sbjct 185 AANCEILVDYISFLKLADDSEVVERYQQLITNTFVECNMLMRWCPAPNCSHAVKAVCAEP 244
Query 264 AGVSCSDSDSFC 275
V C FC
Sbjct 245 RAVLCKCGHEFC 256
> SPAC328.02
Length=504
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 60/279 (21%), Positives = 106/279 (37%), Gaps = 55/279 (19%)
Query 20 ELDEFIYDDDELMESAVASPQSKGLIPRYHSPESLDRRIDELIRETAALMGLNEAFVGIL 79
E DE DD+ ++ + R S L ++E I + +++ L V L
Sbjct 27 EFDEDAEIDDDGFTVERKRRRAHSVSYRVVSVRDLRASLNEKINQLTSIIDLTREQVLGL 86
Query 80 LRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSAL 139
R F W + L++ + + L+K + +Q RE CE
Sbjct 87 YRYFKWNRERLLERYIDAPEESLQKAGVGLSGSKQ-REVVHHEGTCEI------------ 133
Query 140 KTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTEGPQ 199
C C+ P F+ +C H + A + YLD+ ++EG
Sbjct 134 --------------------CYDEGCL-PF---FSAECDHEFCLACYRQYLDSRISEGES 169
Query 200 RALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSPMCQQAV 259
+CP C ++V++ + ++ R FV + + WCP+P C+ A+
Sbjct 170 VI---QCPEESCTQIVSIQSITKVLDEKSLDRYHRLLDRSFVDDNDHLRWCPAPDCEFAI 226
Query 260 ELVPAGVSCSDSDSFCNFSAAEV-ALVADVTCSCGARFC 297
E C+ + A + ++V VTC+CG +FC
Sbjct 227 E--------------CHVTQASLSSVVPTVTCNCGKQFC 251
> At1g65430
Length=575
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 49/218 (22%), Positives = 76/218 (34%), Gaps = 34/218 (15%)
Query 62 IRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQDREDAPG 121
I + ++ ++ ILLR ++W + EWF D V DA G
Sbjct 67 ISRISTVLSISRNSSAILLRHYNWCVSRVHDEWFADEEKV---------------RDAVG 111
Query 122 TAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRY 181
E + +SA+ +C + + A C H +
Sbjct 112 LLEKPVVDFPTDGENSAVSFW--------------QLDCGICFETFLSDKLHAAACGHPF 157
Query 182 SNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFAREQFV 241
++ W GY+ + +GP L RCP P C V D+ N + + +V
Sbjct 158 CDSCWEGYITTAINDGPG-CLTLRCPDPSCRAAVGQDMINLLAPDKDKQKYTSYFVRSYV 216
Query 242 GKHLAVAWCPSPMCQQAVELVPAG----VSCSDSDSFC 275
+ WCP+P C AV V V+C SFC
Sbjct 217 EDNRKTKWCPAPGCDYAVNFVVGSGNYDVNCRCCYSFC 254
> At3g27710
Length=537
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 50/124 (40%), Gaps = 20/124 (16%)
Query 175 LKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRM 234
++C HR+ N WIG+ + EG + + C C + DV + P R
Sbjct 137 MECGHRFCNDCWIGHFTVKINEGESKRI--LCMAHECKAICDEDVVRKLVSPELADRYDR 194
Query 235 FAREQFVGKHLAVAWCPS-PMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCG 293
F E +V + V WCPS P C A+ + G V +V CSCG
Sbjct 195 FLIESYVEDNNMVKWCPSKPHCGSAIRKIEDGHD-----------------VVEVGCSCG 237
Query 294 ARFC 297
+FC
Sbjct 238 LQFC 241
> CE27737
Length=465
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 59/251 (23%), Positives = 93/251 (37%), Gaps = 65/251 (25%)
Query 50 SPESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFN--DSAAVLRKGRL 107
+ +SL+ + + I + A++ + ILL K+ W ++L++ ++ D+ L +
Sbjct 54 NHDSLEAEMKKTITDVQAVLQVKTGVCRILLHKYKWNKESLLERFYEHPDTTTFLIDAHV 113
Query 108 PPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVV 167
P R Q+R A G AEC+ C S
Sbjct 114 IP--RRQERLPA-GDAECD---------------ICCS---------------------- 133
Query 168 PLTETFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMD-VWNRFRGP 226
L E L C+HR W YL + Q ++ C P C L+ + V P
Sbjct 134 -LGELSGLSCNHRACTQCWKAYLTNKIANNAQSEIE--CMAPNCKLLIEDEKVMFYITDP 190
Query 227 SAVARLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVA 286
+ +A R +V + + WCP C +AV + S E LV
Sbjct 191 TVIATYRKLIVASYVETNRLLKWCPGIDCGKAVRV----------------SHWEPRLVV 234
Query 287 DVTCSCGARFC 297
CSCG+RFC
Sbjct 235 ---CSCGSRFC 242
> CE08415
Length=491
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 65/269 (24%), Positives = 106/269 (39%), Gaps = 53/269 (19%)
Query 22 DEFIYDDDELM---ESAVASPQSKGLIPRYHSPESLDRRIDELIRETAALMGLNEAFVGI 78
DE + DDD + +SA K +SL+ + + I E A++ + I
Sbjct 21 DECLSDDDGIARHDQSASDYLNKKDKDNEVLDHDSLEAEMKKAISEVEAVLQVKTGVCRI 80
Query 79 LLRKFDWRSDALIQEWFN--DSAAVLRKGRLPPVVREQDREDAPGTAECESAPAEPEASD 136
LL K+ W ++L++ ++ D+ A L ++ P R+Q+ A G AEC+
Sbjct 81 LLHKYKWNKESLLERFYEHPDTIAFLIDAQVIP--RQQEVIPA-GDAECD---------- 127
Query 137 SALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTE 196
C S+D E L C+HR W YL +
Sbjct 128 -----ICCSMD-----------------------ELSGLSCNHRACAECWQAYLTNKIVS 159
Query 197 GPQRALDARCPFPGCLELVTMD-VWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSPMC 255
Q ++ C P C L+ + V + P+ V++ R +V + + WCP C
Sbjct 160 DAQSEIE--CMAPNCKLLIEDEKVLSYISDPTMVSKYRKLMVASYVEINCLLRWCPGIDC 217
Query 256 QQAVELV---PAGVSCSDSDSFCNFSAAE 281
+AV++ P V CS FC FS +
Sbjct 218 GKAVKVSHWEPRLVVCSCGTCFC-FSCGQ 245
> 7291395
Length=509
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 45/217 (20%), Positives = 87/217 (40%), Gaps = 25/217 (11%)
Query 52 ESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRL-PPV 110
E +++ ++E + + ++ + + +LL + W + A+++++ D+ A+L R+ PP
Sbjct 53 EDIEKLLNERVEKLNTILQITPSLAKVLLLEHQWNNVAVVEKYRQDANALLVTARIKPPS 112
Query 111 VREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLT 170
V D A S KT ++ + + CPV A
Sbjct 113 VAVTDTASTSAAAASAQL---LRLGSSGYKTTASATPQYRSQ------MCPVCASSQLGD 163
Query 171 ETFALKCSHRYSNAAWIGYLDALVTEGPQRAL-------DARCPFPGCLELVTMDVWNRF 223
+ ++L C H + W Y + + +G + + R P L LVT
Sbjct 164 KFYSLACGHSFCKDCWTIYFETQIFQGISTQIGCMAQMCNVRVPEDLVLTLVTR------ 217
Query 224 RGPSAVARLRMFAREQFVGKHLAVAWCPSPMCQQAVE 260
P + + FA + +V H + +CP P CQ V+
Sbjct 218 --PVMRDKYQQFAFKDYVKSHPELRFCPGPNCQIIVQ 252
> CE25598
Length=485
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 65/284 (22%), Positives = 106/284 (37%), Gaps = 57/284 (20%)
Query 17 EGCELDEFIYDDDELMESAVASPQSKGLIPRYHSPESLDRRIDELIRETAALMGLNEAFV 76
E +LDE DD + + Q K P +L+ + + I E A++ +
Sbjct 10 ENNDLDEEFSDDMDQQSGSSGESQGKANY-EILDPTALESDMSKTISEVQAILQVEPGIC 68
Query 77 GILLRKFDWRSDALIQEWF--NDSAAVLRKGRLPPVVREQDREDAPGTAECESAPAEPEA 134
ILL KF W D L+ +++ +D+ L + ++ P + + A SAP P
Sbjct 69 RILLHKFKWNKDRLLDKFYEHSDTTEFLAEAQVIP------KTSSSEEAAGSSAP--PPG 120
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLDALV 194
D+ EC V + L+ L C+HR + W YL +
Sbjct 121 GDA---------------------ECDVCCSMTRLS---GLACAHRACDECWKAYLTEKI 156
Query 195 TEGPQRALDARCPFPGCLELVTMD-VWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSP 253
+ Q ++ C C L+ + V + P +A R +V + + WCP
Sbjct 157 VDVGQSEIE--CMMMDCKLLIEDEKVMSYITDPFVIAAYRKLIISSYVETNSQLKWCPGA 214
Query 254 MCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCGARFC 297
C +AV+ P SD + C+CG RFC
Sbjct 215 GCGKAVKGEP-----SDREP--------------AVCTCGERFC 239
> Hs6453815
Length=557
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 52/250 (20%), Positives = 89/250 (35%), Gaps = 52/250 (20%)
Query 50 SPESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPP 109
+ E + + + E IRE ++ ILL F+W + L++ +F+ G L
Sbjct 105 TAEQILQHMVECIREVNEVIQNPATITRILLSHFNWDKEKLMERYFD--------GNLEK 156
Query 110 VVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPL 169
+ AEC ++ + T ++ D+ C + P
Sbjct 157 LF-----------AECHVINPSKKSRTRQMNTRSSAQDMP----------CQICYLNYPN 195
Query 170 TETFALKCSHRYSNAAWIGYLDA-LVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSA 228
+ L+C H++ W YL ++ EG + CP GC LV + R S
Sbjct 196 SYFTGLECGHKFCMQCWSEYLTTKIMEEGMGHTIS--CPAHGCDILVDDNTVMRLITDSK 253
Query 229 VA-RLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVAD 287
V + + FV + + WCP+P C V++ +
Sbjct 254 VKLKYQHLITNSFVECNRLLKWCPAPDCHHVVKV-------------------QYPDAKP 294
Query 288 VTCSCGARFC 297
V C CG +FC
Sbjct 295 VRCKCGRQFC 304
> CE08416
Length=497
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 55/247 (22%), Positives = 95/247 (38%), Gaps = 65/247 (26%)
Query 54 LDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFN--DSAAVLRKGRLPPVV 111
L+ ++ I + A++ ++ ILL K+ W ++L++ + D+ A L ++ P
Sbjct 61 LEAEMNTTIADVQAVLQVDPGVCRILLHKYKWNKESLLERLYEHPDTIAFLIDAQVIP-- 118
Query 112 REQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTE 171
R+Q+ A G AEC+ C S+D E
Sbjct 119 RQQEVIPA-GDAECD---------------ICCSMD-----------------------E 139
Query 172 TFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMD-VWNRFRGPSAVA 230
L C+HR W YL + Q ++ C P C L+ + V + P+ +A
Sbjct 140 LSGLSCNHRACAECWQAYLTNKIVSDAQSEIE--CMAPNCKLLIEDEKVLAYIKDPTIIA 197
Query 231 RLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTC 290
+ R ++ + + WCP C + V++ S E LV C
Sbjct 198 KYRKMMVASYIEINALLKWCPGVDCGRTVKV----------------SHGEPRLVV---C 238
Query 291 SCGARFC 297
+CG+RFC
Sbjct 239 TCGSRFC 245
> At2g31510
Length=565
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 54/230 (23%), Positives = 80/230 (34%), Gaps = 60/230 (26%)
Query 52 ESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVV 111
E + R + I + ++ + E ILLR F W + EWF D V +
Sbjct 66 EDIRRHQMDNIERVSVVLSITEVEASILLRHFHWSVGRVHDEWFADEERVRK-------- 117
Query 112 REQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTE 171
T + P + DS L C + P +
Sbjct 118 ----------TVGILESHVVPPSDDSELT-------------------CGICFDSYPPEK 148
Query 172 TFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDV------------ 219
++ C H + W GY+ + +GP L RCP P CL V D+
Sbjct 149 IASVSCGHPFCTTCWTGYISTTINDGPG-CLMLRCPDPSCLAAVGHDMVDKLASEDEKEK 207
Query 220 WNRFRGPSAVARLR------MF---AREQFVGKHLAVAWCPSPMCQQAVE 260
+NR+ S + R MF AR F G LA + P P C++ +E
Sbjct 208 YNRYFLRSYIEDNRKGLEIMMFPACARLAFAGMILANSK-PCPRCKRPIE 256
> 7293430
Length=503
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 51/223 (22%), Positives = 79/223 (35%), Gaps = 41/223 (18%)
Query 60 ELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRK--GRLPPVVREQDRE 117
E+I E L+ L ILL F W + L++++F+D+ K + P + +
Sbjct 63 EIIDEANLLLKLPTPTTRILLNHFKWDKEKLLEKYFDDNTDEFFKCAHVINPFNATEAIK 122
Query 118 DAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALKC 177
++CE EC + +P L+C
Sbjct 123 QKTSRSQCE--------------------------------ECEICFSQLPPDSMAGLEC 150
Query 178 SHRYSNAAWIGYLDA-LVTEGPQRALDARCPFPGCLELV-TMDVWNRFRGPSAVARLRMF 235
HR+ W YL +V EG + + C GC LV + V N + +
Sbjct 151 GHRFCMPCWHEYLSTKIVAEGLGQTIS--CAAHGCDILVDDVTVANLVTDARVRVKYQQL 208
Query 236 AREQFVGKHLAVAWCPSPMCQQAVEL---VPAGVSCSDSDSFC 275
FV + + WCPS C AV++ P V C FC
Sbjct 209 ITNSFVECNQLLRWCPSVDCTYAVKVPYAEPRRVHCKCGHVFC 251
> At4g34370
Length=594
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/142 (23%), Positives = 53/142 (37%), Gaps = 25/142 (17%)
Query 160 CPVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDV 219
C V +P + C H + N W + + EG + + RC C + D+
Sbjct 123 CDVCMEDLPGDHMTRMDCGHCFCNNCWTEHFTVQINEGQSKRI--RCMAHQCNAICDEDI 180
Query 220 WNRF---RGPSAVARLRMFAREQFVGKHLAVAWCPS-PMCQQAVELVPAGVSCSDSDSFC 275
+ P A+ + E ++ + V WCPS P C A+ ++ D C
Sbjct 181 VRSLVSKKRPDLAAKFDRYLLESYIEDNRMVKWCPSTPHCGNAIR--------AEDDKLC 232
Query 276 NFSAAEVALVADVTCSCGARFC 297
+V CSCG +FC
Sbjct 233 -----------EVECSCGLQFC 243
> YKR017c
Length=551
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 55/258 (21%), Positives = 94/258 (36%), Gaps = 53/258 (20%)
Query 41 SKGLIPRYHSPESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAA 100
+KG+ R + +R+D L + + A + ILL+ +DW + L++ W
Sbjct 96 TKGIFER------MLQRVDHL----QPIFAIPSADILILLQHYDWNEERLLEVWTEKMDE 145
Query 101 VLRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFEC 160
+L + L + D ++ E + F C
Sbjct 146 LLVELGLSTTANIKKDNDYNSHF--------------------------REVEFKNDFTC 179
Query 161 PVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVW 220
+ C TETFAL+C H Y + Y+ + EG + L L D+
Sbjct 180 -IICCDKKDTETFALECGHEYCINCYRHYIKDKLHEG-----NIITCMDCSLALKNEDI- 232
Query 221 NRFRGPSAVARLRMFAREQFVGKH-LAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSA 279
++ G + ++L + + FV KH WCP C+ V L D+ S ++
Sbjct 233 DKVMGHPSSSKLMDSSIKSFVQKHNRNYKWCPFADCKSIVHL-------RDTSSLPEYTR 285
Query 280 AEVALVADVTCSCGARFC 297
+ V C+ RFC
Sbjct 286 LHYSPF--VKCNSFHRFC 301
> At2g16090
Length=518
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 38/99 (38%), Gaps = 6/99 (6%)
Query 167 VPLTETFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRF--- 223
VP + + C H + N W G+ + EG + + C C + DV
Sbjct 120 VPGYQLTRMDCGHSFCNNCWTGHFTVKINEGQSKRI--ICMAHKCNAICDEDVVRALVSK 177
Query 224 RGPSAVARLRMFAREQFVGKHLAVAWCPS-PMCQQAVEL 261
P + F E ++ + V WCPS P C A+ +
Sbjct 178 SQPDLAEKFDRFLLESYIEDNKMVKWCPSTPHCGNAIRV 216
> At5g63750
Length=536
Score = 38.9 bits (89), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 47/256 (18%), Positives = 83/256 (32%), Gaps = 63/256 (24%)
Query 52 ESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVV 111
+ L + + I E + + L++ +LL W S + + ++ VL + L PVV
Sbjct 15 DQLKGNMKKQIAEISEIFSLSKPDATVLLMFLRWDSHEVSEFLVENNEKVLSESGLKPVV 74
Query 112 REQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTE 171
+ + +D + C + D + TP
Sbjct 75 VDPN-QDLYKISSCGICFKTCDDGDYLISTPF---------------------------- 105
Query 172 TFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVAR 231
CSH + + W YL+ + CP C V GP + +
Sbjct 106 -----CSHMFCKSCWRKYLEKNFYLVEKTQTRISCPHGACQAAV---------GPDTIQK 151
Query 232 LRMFAREQFV----------GKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAE 281
L + +E +V K L + +CP+ C +E +D + +
Sbjct 152 LTVCDQEMYVEYILRSYIEGNKVLEIKYCPAQDCNYVIEFHQKNHDGADQEDY------- 204
Query 282 VALVADVTCSCGARFC 297
+V C CG FC
Sbjct 205 ---GFNVVCLCGHIFC 217
> Hs4757762
Length=474
Score = 37.4 bits (85), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 48/125 (38%), Gaps = 6/125 (4%)
Query 157 MFECPVTACVVPLTE-TFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELV 215
+F C + C +E + L+C H Y A Y + + +G + L+ CP P C +
Sbjct 217 LFLCSICFCEKLGSECMYFLECRHVYCKACLKDYFEIQIRDGQVQCLN--CPEPKCPSVA 274
Query 216 TMDVWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPA---GVSCSDSD 272
T AR + + V +CP P CQ V P G+ S +
Sbjct 275 TPGQVKELVEAELFARYDRLLLQSSLDLMADVVYCPRPCCQLPVMQEPGCTMGICSSCNF 334
Query 273 SFCNF 277
+FC
Sbjct 335 AFCTL 339
> CE27704
Length=793
Score = 35.4 bits (80), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 59/132 (44%), Gaps = 17/132 (12%)
Query 131 EPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALK-CSHRYSNAAWIGY 189
E + S S TP T +V KK + M ECP+ A +P + LK C HR A Y
Sbjct 57 EKQLSQSLPDTPKTPSEVGKKGKG-KMKECPLCAAKMPGSAFPKLKGCQHRSCRACLRQY 115
Query 190 LDALVTEGPQRALDARCP------FPGCLELVTMDVWNRFRGPSAVARLRMFAREQFVGK 243
++ +TE ++ CP P ++++ D+ P+ + + F+ +++
Sbjct 116 VELSITEN---RVEVPCPECSSYLHPNDIKMLIGDI------PTLIEKYEAFSLRRYLMT 166
Query 244 HLAVAWCPSPMC 255
WCP+P C
Sbjct 167 EADARWCPAPDC 178
> CE27705
Length=796
Score = 35.4 bits (80), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 59/132 (44%), Gaps = 17/132 (12%)
Query 131 EPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPLTETFALK-CSHRYSNAAWIGY 189
E + S S TP T +V KK + M ECP+ A +P + LK C HR A Y
Sbjct 57 EKQLSQSLPDTPKTPSEVGKKGKG-KMKECPLCAAKMPGSAFPKLKGCQHRSCRACLRQY 115
Query 190 LDALVTEGPQRALDARCP------FPGCLELVTMDVWNRFRGPSAVARLRMFAREQFVGK 243
++ +TE ++ CP P ++++ D+ P+ + + F+ +++
Sbjct 116 VELSITEN---RVEVPCPECSSYLHPNDIKMLIGDI------PTLIEKYEAFSLRRYLMT 166
Query 244 HLAVAWCPSPMC 255
WCP+P C
Sbjct 167 EADARWCPAPDC 178
> CE02616
Length=214
Score = 35.0 bits (79), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 8/94 (8%)
Query 72 NEAFV-----GILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQDREDAPGTAECE 126
NE F+ ++LR +A ++ F+ L+K R P E P T +
Sbjct 51 NERFLPGKKKNLMLRVIQAEPNAFLEATFDRD---LKKKRNPKASTTNVCESTPATIDLV 107
Query 127 SAPAEPEASDSALKTPCTSVDVSKKEEAGDMFEC 160
+ PA P SDS ++ P S ++ + D+ EC
Sbjct 108 TPPASPRRSDSYMQFPSNSRELDEPRSTDDIPEC 141
> CE02605
Length=585
Score = 34.7 bits (78), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 8/94 (8%)
Query 72 NEAFV-----GILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQDREDAPGTAECE 126
NE F+ ++LR +A ++ F+ L+K R P E P T +
Sbjct 51 NERFLPGKKKNLMLRVIQAEPNAFLEATFDRD---LKKKRNPKASTTNVCESTPATIDLV 107
Query 127 SAPAEPEASDSALKTPCTSVDVSKKEEAGDMFEC 160
+ PA P SDS ++ P S ++ + D+ EC
Sbjct 108 TPPASPRRSDSYMQFPSNSRELDEPRSTDDIPEC 141
> CE25342
Length=382
Score = 34.3 bits (77), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 54/121 (44%), Gaps = 8/121 (6%)
Query 151 KEEAGDMFECPVTACVVPLTETFAL-KCSHRYSNAAWIGYLDALVTEGPQRALDARCPFP 209
+EE+G M EC + +VP L C H Y Y++ + ++ CP
Sbjct 32 QEESGAMKECELCCEMVPAGAFCQLINCRHVYCRICIRKYMELSIL---GNRVEIPCP-G 87
Query 210 GCLELVTMDVWNRFRGPSA--VARLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAGVS 267
GC ++ + R+ P+ +++ F+ + + V WCP+P C AV +VP G
Sbjct 88 GCPAVIHPNDVTRYLLPNTDLISKYESFSIRMALCRIQDVRWCPAPDCGFAV-IVPNGQK 146
Query 268 C 268
C
Sbjct 147 C 147
> HsM4885113
Length=1288
Score = 33.1 bits (74), Expect = 0.71, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 103 RKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKE 152
RKG P R ++ P + + S+ AEPE ++P T+ + + +E
Sbjct 3 RKGTQPSTARRREEGPPPPSPDGASSDAEPEPPSGRTESPATAAETASEE 52
> Hs21361252
Length=1288
Score = 33.1 bits (74), Expect = 0.71, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 103 RKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKE 152
RKG P R ++ P + + S+ AEPE ++P T+ + + +E
Sbjct 3 RKGTQPSTARRREEGPPPPSPDGASSDAEPEPPSGRTESPATAAETASEE 52
> At5g63730
Length=506
Score = 32.7 bits (73), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 28/122 (22%), Positives = 39/122 (31%), Gaps = 14/122 (11%)
Query 177 CSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLRMFA 236
CSH++ + W YL+ + CP C V D + R
Sbjct 106 CSHKFDSKYWREYLEKNFYYVEKIQTTISCPDQDCRSAVGPDTIEKLTVRDQEMYERYIW 165
Query 237 REQFVG-KHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCGAR 295
R G K L + CP+ C +E D D + +V C CG
Sbjct 166 RSYIEGNKVLMIKQCPARNCDYVIEFHQEN---DDDDEY----------SLNVVCICGHI 212
Query 296 FC 297
FC
Sbjct 213 FC 214
> CE28790
Length=557
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 0/59 (0%)
Query 102 LRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFEC 160
L+K R P E P T + + PA P SDS ++ P S ++ + D+ EC
Sbjct 31 LKKKRNPKASTTNVCESTPATIDLVTPPASPRRSDSYMQFPSNSRELDEPRSTDDIPEC 89
> At5g36860
Length=1204
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 41/88 (46%), Gaps = 18/88 (20%)
Query 78 ILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQD---REDAPGTAECESAPAEPEA 134
+L+ W+ D EW A V ++ R PP + E D + +APG +P EP
Sbjct 386 LLIDGHQWQED----EWVGGYARVPKQSR-PPQLEETDVKRKRNAPG-----PSPKEP-- 433
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPV 162
A+K + +D K+E A D F PV
Sbjct 434 ---AMKNQKSEMDYDKEETAEDCFGEPV 458
> CE03451
Length=815
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 33/73 (45%), Gaps = 0/73 (0%)
Query 92 QEWFNDSAAVLRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKK 151
+++F+ L+K R P E P T + + PA P SDS ++ P S ++ +
Sbjct 104 EKYFSAFDRDLKKKRNPKASATNVCESTPATIDLVTPPASPRRSDSYMQFPSNSRELDEP 163
Query 152 EEAGDMFECPVTA 164
D+ EC A
Sbjct 164 RSTDDIPECDSIA 176
> CE13583
Length=488
Score = 32.3 bits (72), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 47/233 (20%), Positives = 82/233 (35%), Gaps = 42/233 (18%)
Query 50 SPESLDRRIDELIRETAALMGLNEAFVGILLRKFDWRSDALIQEWFNDSAAVLRKGRLPP 109
S ++R + + + + +NE F ILL+ W D + + ND LRK +
Sbjct 50 SVNQVERVFIDGVNSLVSRISINEKFARILLQANHWDVDKIARLVRNDRNDFLRKCHI-- 107
Query 110 VVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFECPVTACVVPL 169
A EP+ S+ ++ C V A +
Sbjct 108 -----------------DAKPEPKRKLSSTQSVLAKG------------YCSVCA-MDGY 137
Query 170 TETFALKCSHRYSNAAWIGYLDALVTEGPQ---RALDARCPFPGCLELVTMDVWNRFRGP 226
TE L C H + W ++++ ++EG +++ C E V + N P
Sbjct 138 TELPHLTCGHCFCEHCWKSHVESRLSEGVASRIECMESECEVYAPSEFVLSIIKN---SP 194
Query 227 SAVARLRMFAREQFVGKHLAVAWCPS---PMCQQAVELVPAGVSCSD-SDSFC 275
+ F V H + +C P+ ++ E+ P V+C SFC
Sbjct 195 VIKLKYERFLLRDMVNSHPHLKFCVGNECPVIIRSTEVKPKRVTCMQCHTSFC 247
> At3g27720
Length=493
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 27/64 (42%), Gaps = 18/64 (28%)
Query 235 FAREQFVGKHLAVAWCPS-PMCQQAVELVPAGVSCSDSDSFCNFSAAEVALVADVTCSCG 293
F E +V + V WCPS P C A+ + D D V +V CSCG
Sbjct 155 FLIESYVEDNNMVKWCPSTPHCGNAIRNIK-----DDGD------------VDEVECSCG 197
Query 294 ARFC 297
+FC
Sbjct 198 LQFC 201
> At1g34740
Length=1383
Score = 31.2 bits (69), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 41/88 (46%), Gaps = 18/88 (20%)
Query 78 ILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQD---REDAPGTAECESAPAEPEA 134
+L+ W+ D EW A V ++ R PP + E D + +APG +P EP
Sbjct 366 LLIDGHQWQED----EWVGGYARVPKQSR-PPQLEETDVKRKRNAPG-----PSPKEP-- 413
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPV 162
A+K + +D K+E A D F PV
Sbjct 414 ---AMKKQKSEMDCDKEETAEDCFGEPV 438
> Hs22042144
Length=1538
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 42/123 (34%), Gaps = 2/123 (1%)
Query 92 QEWFNDSAAVLRKGRLP-PVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSK 150
Q+ + + R P P ++ R APGT + PA DS TP T D S+
Sbjct 809 QDSRTPTPGTQQDSRTPTPGTQQDSRTPAPGTQQDSRTPAPGSQQDSRTPTPGTQQD-SR 867
Query 151 KEEAGDMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLDALVTEGPQRALDARCPFPG 210
G + A L S + S G T P D+R P PG
Sbjct 868 TPAPGSQLDSRTPAPGSQLDSRTPAPGSQQDSRTPAPGTQQDSRTPAPGTQQDSRTPAPG 927
Query 211 CLE 213
+
Sbjct 928 SQQ 930
> At1g25886
Length=1201
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 41/88 (46%), Gaps = 18/88 (20%)
Query 78 ILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQD---REDAPGTAECESAPAEPEA 134
+L+ W+ D EW A V ++ R PP + E D + +APG +P EP
Sbjct 353 LLIDGHQWQED----EWVGGYARVPKQSR-PPQLEETDVKRKRNAPG-----PSPKEP-- 400
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPV 162
A+K + +D K+E A D F PV
Sbjct 401 ---AMKKQKSEMDCDKEETAEDCFGEPV 425
> At4g05280
Length=1312
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 41/88 (46%), Gaps = 18/88 (20%)
Query 78 ILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQD---REDAPGTAECESAPAEPEA 134
+L+ W+ D EW A V ++ R PP + E D + +APG +P EP
Sbjct 332 LLIDGHQWQED----EWVGGYARVPKQLR-PPQLEETDVKRKRNAPG-----PSPKEP-- 379
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPV 162
A+K + +D K+E A D F PV
Sbjct 380 ---AMKKQISEMDCDKEENAEDCFGEPV 404
> Hs4504747
Length=1051
Score = 30.8 bits (68), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 39/94 (41%), Gaps = 15/94 (15%)
Query 137 SALKTPCTSVDVSKKEEAG-----DMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLD 191
+A K C ++++ K + G DM+ A P L C+HRY+ W G D
Sbjct 97 TAHKDDCERMNITVKNDPGHHIIEDMWLGVTVASQGPAGR--VLVCAHRYTQVLWSGSED 154
Query 192 ALVTEGPQRALDARCPFPGC-LELVTMDVWNRFR 224
QR + +C G LEL + D W +
Sbjct 155 -------QRRMVGKCYVRGNDLELDSSDDWQTYH 181
> Hs6006011
Length=1066
Score = 30.4 bits (67), Expect = 4.1, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 39/93 (41%), Gaps = 15/93 (16%)
Query 137 SALKTPCTSVDVSKKEEAG-----DMFECPVTACVVPLTETFALKCSHRYSNAAWIGYLD 191
+A K C ++++ K + G DM+ A P L C+HRY+ W G D
Sbjct 97 TAHKDDCERMNITVKNDPGHHIIEDMWLGVTVASQGPAGRV--LVCAHRYTQVLWSGSED 154
Query 192 ALVTEGPQRALDARCPFPGC-LELVTMDVWNRF 223
QR + +C G LEL + D W +
Sbjct 155 -------QRRMVGKCYVRGNDLELDSSDDWQTY 180
> At5g63760
Length=503
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 29/133 (21%), Positives = 46/133 (34%), Gaps = 37/133 (27%)
Query 177 CSHRYSNAAWIGYLDALVTEGPQRALDARCPFPGCLELVTMDVWNRFRGPSAVARLR--- 233
CSH++ A W YL + CP C V GP V +L
Sbjct 54 CSHKFCKACWSKYLKKNFFSVEKNHTAISCPDRDCRAAV---------GPETVEKLTVRD 104
Query 234 --MF-------AREQFVGKHLAVAWCPSPMCQQAVELVPAGVSCSDSDSFCNFSAAEVAL 284
M+ RE+++G L + CP+ C +E + ++ +
Sbjct 105 QAMYELYILKSYREKYLGWKLKL--CPARGCNYVIEF--------------HLASEDEEH 148
Query 285 VADVTCSCGARFC 297
++ C CG FC
Sbjct 149 SLNIVCLCGHIFC 161
> At4g08880
Length=1175
Score = 29.6 bits (65), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 41/88 (46%), Gaps = 18/88 (20%)
Query 78 ILLRKFDWRSDALIQEWFNDSAAVLRKGRLPPVVREQD---REDAPGTAECESAPAEPEA 134
+L+ W+ D +W A V ++ R PP + E D + +APG + P EP
Sbjct 250 LLIDGHQWQED----DWVGGYARVPKQVR-PPQLEETDVKRKRNAPGLS-----PKEP-- 297
Query 135 SDSALKTPCTSVDVSKKEEAGDMFECPV 162
A+K + +D K+E A D F PV
Sbjct 298 ---AMKIQKSEMDCDKEENAEDCFGEPV 322
> CE29101
Length=796
Score = 29.6 bits (65), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 26/59 (44%), Gaps = 0/59 (0%)
Query 102 LRKGRLPPVVREQDREDAPGTAECESAPAEPEASDSALKTPCTSVDVSKKEEAGDMFEC 160
L+K R P E P T + + PA P SD ++ P S ++ + D+ EC
Sbjct 31 LKKKRNPKASTTNVCESTPATIDLVTPPASPRRSDLYMQFPSNSRELDEPRSTDDIPEC 89
> Hs7661956
Length=292
Score = 29.3 bits (64), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 51/122 (41%), Gaps = 24/122 (19%)
Query 161 PVTACVVPLTE------TFALKCSHRYSNAAWIGYLDALVTEGPQRAL---DARCPFPGC 211
P+ +C + L E T +C + Y++ L+ EG + A+ DA CP G
Sbjct 16 PLVSCKLCLGEYPVEQMTTIAQCQCIFCTLCLKQYVELLIKEGLETAISCPDAACPKQGH 75
Query 212 LE------LVTMDVWNRFRGPSAVARLRMFAREQFVGKHLAVAWCPSPMCQQAVELVPAG 265
L+ +V ++ R++ +L+ F RE WCP+ CQ +L G
Sbjct 76 LQENEIECMVAAEIMQRYK------KLQ-FEREVLFDP--CRTWCPASTCQAVCQLQDVG 126
Query 266 VS 267
+
Sbjct 127 LQ 128
Lambda K H
0.320 0.133 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6672668814
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40