bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1604_orf3
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
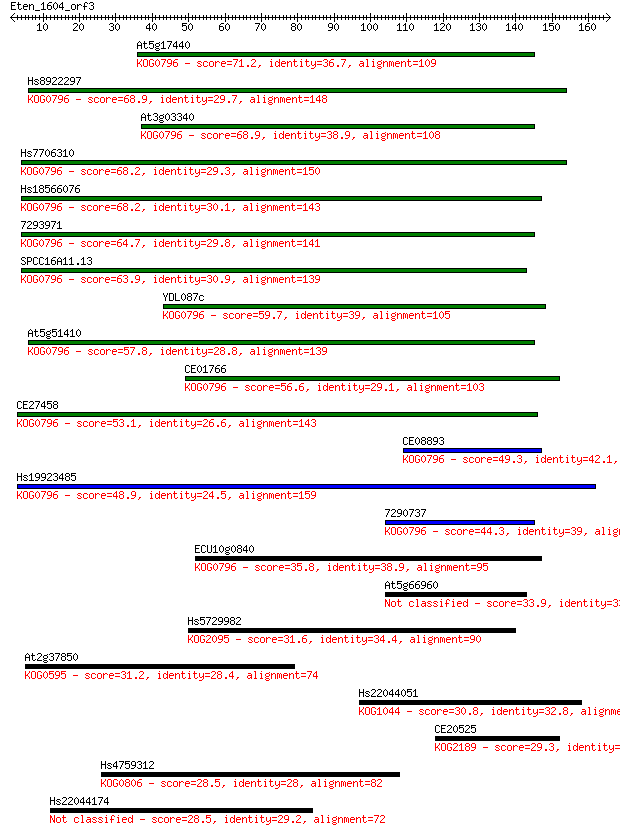
At5g17440 71.2 9e-13
Hs8922297 68.9 4e-12
At3g03340 68.9 5e-12
Hs7706310 68.2 7e-12
Hs18566076 68.2 8e-12
7293971 64.7 8e-11
SPCC16A11.13 63.9 1e-10
YDL087c 59.7 3e-09
At5g51410 57.8 9e-09
CE01766 56.6 2e-08
CE27458 53.1 3e-07
CE08893 49.3 3e-06
Hs19923485 48.9 5e-06
7290737 44.3 1e-04
ECU10g0840 35.8 0.037
At5g66960 33.9 0.16
Hs5729982 31.6 0.80
At2g37850 31.2 0.96
Hs22044051 30.8 1.4
CE20525 29.3 3.7
Hs4759312 28.5 6.7
Hs22044174 28.5 6.9
> At5g17440
Length=381
Score = 71.2 bits (173), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 67/117 (57%), Gaps = 8/117 (6%)
Query 36 SLPGVENVKSVHPEVLRLDQAIAQKFKEMEDLGKLGRIDQSMGVSLELTSLQTRRAK--- 92
++P + ++ P R + I +K K+ E+LG+ G +D++ E +L+ A+
Sbjct 166 AVPAQQPAATLPPPDPRTQEMINEKLKKAEELGEQGMVDEAQKALEEAEALKKLTARQEP 225
Query 93 ----LLFRNAEGQLT-QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDAL 144
+ A+ ++T Q+LR CD+CGA LS D+DRRL +HF G++HLG +RD L
Sbjct 226 VVDSTKYTAADVRITDQKLRLCDICGAFLSVYDSDRRLADHFGGKLHLGYMLIRDKL 282
> Hs8922297
Length=325
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 82/153 (53%), Gaps = 5/153 (3%)
Query 6 FEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKEME 65
FE A+ + + E D++ + A+ R+ V + +V L++ I + + E
Sbjct 82 FELDAMDHLESFIAECDRRTELAKKRLAETQEEISAEVSAKAEKVHELNEEIGKLLAKAE 141
Query 66 DLGKLGRIDQSMGVSLELTSLQTRR--AKLLFRN---AEGQLTQRLRPCDVCGALLSAND 120
LG G +D+S + +E+ ++ ++ A+ +RN A Q+LR C+VC A L +D
Sbjct 142 QLGAEGNVDESQKILMEVEKVRAKKKEAEEEYRNSMPASSFQQQKLRVCEVCSAYLGLHD 201
Query 121 TDRRLDEHFSGRVHLGLQKLRDALKKNLEYIAQ 153
DRRL +HF G++HLG ++R+ L + + +A+
Sbjct 202 NDRRLADHFGGKLHLGFIQIREKLDQLRKTVAE 234
> At3g03340
Length=385
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 62/116 (53%), Gaps = 8/116 (6%)
Query 37 LPGVENVKSVHPEVLRLDQAIAQKFKEMEDLGKLGRIDQSMGVSLE---LTSLQTRRA-- 91
+P + P R + I +K K+ EDLG+ G +D++ E L L RR
Sbjct 165 VPAQPPSSELPPPDPRTQEMINEKLKKAEDLGEQGMVDEAQKALEEAEALKKLTVRREPP 224
Query 92 --KLLFRNAEGQLT-QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDAL 144
+ + ++T Q+LR CD+CGA LS D+DRRL +HF G++HLG +RD L
Sbjct 225 ADSTKYTAVDVRITDQKLRLCDICGAFLSVYDSDRRLADHFGGKLHLGYMLVRDKL 280
> Hs7706310
Length=402
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 82/155 (52%), Gaps = 5/155 (3%)
Query 4 YGFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKE 63
+ FE A+ ++ + + D++ + A+ R+ V + V L++ I + +
Sbjct 80 FFFELDAMDHLQSFIADCDRRTEVAKKRLAETQEEISAEVAAKAERVHELNEEIGKLLAK 139
Query 64 MEDLGKLGRIDQSMGV--SLELTSLQTRRAKLLFRN---AEGQLTQRLRPCDVCGALLSA 118
+E LG G +++S V +E + R A+ ++RN A Q+LR C+VC A L
Sbjct 140 VEQLGAEGNVEESQKVMDEVEKARAKKREAEEVYRNSMPASSFQQQKLRVCEVCSAYLGL 199
Query 119 NDTDRRLDEHFSGRVHLGLQKLRDALKKNLEYIAQ 153
+D DRRL +HF G++HLG ++R+ L++ +A+
Sbjct 200 HDNDRRLADHFGGKLHLGFIEIREKLEELKRVVAE 234
> Hs18566076
Length=392
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 79/148 (53%), Gaps = 5/148 (3%)
Query 4 YGFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKE 63
+ FE A+ ++ + + D++ + A+ R+ V + V L++ I + +
Sbjct 80 FFFELDAMDHLQSFIADCDRRTEVAKKRLAETQEEISAEVAAKAERVHELNEEIGKLLAK 139
Query 64 MEDLGKLGRIDQSMGV--SLELTSLQTRRAKLLFRN---AEGQLTQRLRPCDVCGALLSA 118
+E LG G +++S V +E + R A+ ++RN A Q+LR C+VC A L
Sbjct 140 VEQLGAEGNVEESQKVMDEVEKARAKKREAEEVYRNSMPASSFQQQKLRVCEVCSAYLGL 199
Query 119 NDTDRRLDEHFSGRVHLGLQKLRDALKK 146
+D DRRL +HF G++HLG ++R+ L++
Sbjct 200 HDNDRRLADHFGGKLHLGFIEIREKLEE 227
> 7293971
Length=420
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 76/146 (52%), Gaps = 5/146 (3%)
Query 4 YGFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKE 63
Y ++ A++ +A + + D++ +A+ R+ V V L + I +K +
Sbjct 85 YYYDIEAMEHLQAFIADCDRRTDSAKQRLKETQEELTAEVAEKANAVHGLAEEIGKKLAK 144
Query 64 MEDLGKLGRIDQSMGVSLELTSLQTRR--AKLLFRNAEGQLT---QRLRPCDVCGALLSA 118
E LG+ G ++ SM + E+ L+ ++ A+ +R + T Q+LR C+VC A L
Sbjct 145 AEALGEAGEVEDSMELMKEIEELRAKKIKAEHEYRTSMPASTYQQQKLRVCEVCSAYLGI 204
Query 119 NDTDRRLDEHFSGRVHLGLQKLRDAL 144
+D D RL +HF G++HLG +R+ L
Sbjct 205 HDNDIRLADHFGGKLHLGFLTIREKL 230
> SPCC16A11.13
Length=264
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 75/147 (51%), Gaps = 9/147 (6%)
Query 4 YGFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKE 63
YG+E ++ V++ +K++ A+ R + + E + + +++ D +I E
Sbjct 78 YGYEWDYLEDLERHVDDCNKRIDIAEARRE-KTKEEEERIDELMRDIIHTDHSIEVIITE 136
Query 64 MEDLGKLGRIDQSMGVSLELTSLQTRRAKLL--------FRNAEGQLTQRLRPCDVCGAL 115
ME L K ++ ++ +EL L+T R +L + Q+L+ CD+C A
Sbjct 137 MEALAKRKLVNDAVKHFIELNRLKTYRKELYDEVISMNEIPSQASTTHQKLQVCDICSAY 196
Query 116 LSANDTDRRLDEHFSGRVHLGLQKLRD 142
LS D DRRL +HFSG++HLG LR+
Sbjct 197 LSRLDNDRRLADHFSGKMHLGYAMLRN 223
> YDL087c
Length=261
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 43 VKSVHPEVLRLDQAIAQKFKEMEDLGKLGRIDQSMGVSLELTSLQTRRAKLLFR------ 96
++ V E+ LD I +E++ L + + M S++L L ++R ++ R
Sbjct 129 IQQVTEELDVLDVRIGLMGQEIDSLIRADEVSMGMLQSVKLQELISKRKEVAKRVRNITE 188
Query 97 NAEGQLTQRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLR---DALKKN 147
N Q+L+ C+VCGA LS DTDRRL +HF G++HLG K+R D L KN
Sbjct 189 NVGQSAQQKLQVCEVCGAYLSRLDTDRRLADHFLGKIHLGYVKMREDYDRLMKN 242
> At5g51410
Length=334
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 40/147 (27%), Positives = 73/147 (49%), Gaps = 10/147 (6%)
Query 6 FEELAVQLCRALVEEADKKVQTAQMR----VDCASLPGVENVKSVHPEVLRLDQAIAQKF 61
FE Q C LV + D+KV+ + R V+ P + K+ VL ++ +
Sbjct 85 FEAELAQFCEKLVNDLDRKVRRGRERLAQEVEPVPPPSLSAEKAEQLSVL--EEKVKNLL 142
Query 62 KEMEDLGKLGRIDQSMGVSLELTSLQTRRAKLLFRNAEGQLT----QRLRPCDVCGALLS 117
+++E LG+ G++D++ + ++ L + LL R + L +++ C+VCG+ L
Sbjct 143 EQVEALGEEGKVDEAEALMRKVEGLNAEKTVLLQRPTDKVLAMAQEKKMALCEVCGSFLV 202
Query 118 ANDTDRRLDEHFSGRVHLGLQKLRDAL 144
AND R H +G+ H+G +RD +
Sbjct 203 ANDAVERTQSHVTGKQHVGYGLVRDFI 229
> CE01766
Length=313
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 61/109 (55%), Gaps = 6/109 (5%)
Query 49 EVLRLDQAIAQKFKEMEDLGKLGRIDQSMGVSLELTSLQTRRAKLLFRNAE------GQL 102
+V +++ IA+ ++E LG G+I++SM + + L+ + ++ E G
Sbjct 125 QVAEIEEKIAKNVDDIEKLGNEGKIEESMKLHKYVEELREKIQEIEDSQTEVKTAGPGSN 184
Query 103 TQRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDALKKNLEYI 151
+ +LR C+ CGA L+ D + R+ +H++G++H+G+ + R+ K E I
Sbjct 185 SAKLRVCEDCGAQLNITDHESRIADHYNGKMHIGMVETRETYLKMKETI 233
> CE27458
Length=339
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/167 (22%), Positives = 81/167 (48%), Gaps = 26/167 (15%)
Query 3 RYGFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFK 62
R GFE+ ++ R + E+ +K+Q + R+ ++ ++L ++Q Q K
Sbjct 79 RMGFEKRFLERIRRIHEDVRRKIQKHEDRLAVTQGESKSAEETFGQKILEIEQRREQLTK 138
Query 63 EMEDL-------GKLGRIDQSMG--------------VSLELTSLQTRRAKLLFRNAEGQ 101
+MEDL G+ G +D + ++ E L++ + + + N E
Sbjct 139 KMEDLMDEAALEGEKGNVDAAQTAVDRADKAKVEVEELTQEAEKLKSEKERAI--NMEEN 196
Query 102 LT---QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDALK 145
+T ++++ C +CG + ND +R+D+H +G++H+ Q++ D +K
Sbjct 197 VTAGNRQMQVCQICGCFMLQNDAPQRVDDHLTGKLHIAYQQISDTIK 243
> CE08893
Length=369
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 109 CDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDALKK 146
C+VCG++L ND +R++EH +G++H G QK+R +++
Sbjct 204 CEVCGSMLIVNDAQQRIEEHLTGKMHTGFQKIRTMIQQ 241
> Hs19923485
Length=432
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 39/176 (22%), Positives = 85/176 (48%), Gaps = 23/176 (13%)
Query 3 RYGFEELAVQLCRALVEEADKKVQTAQMRV--------DCASLPGVENVKSVHPEVLRLD 54
+ G+E ++ ++L+ E +++++ R+ A+ P +N + + ++D
Sbjct 78 KVGYERDFLRYLQSLLAEVERRIRRGHARLALSQNQQSSGAAGPTGKNEEKIQVLTDKID 137
Query 55 QAIAQKFKEMEDLGKLGRIDQSMGVSLELTSLQTRRAKLLFRNAEGQLT------QRLRP 108
+ Q +E+LG G+++++ G+ + L+ R L R+ + +++
Sbjct 138 VLLQQ----IEELGSEGKVEEAQGMMKLVEQLKEERE--LLRSTTSTIESFAAQEKQMEV 191
Query 109 CDVCGALLSANDTDRRLDEHFSGRVHLGLQKLR---DALKKNLEYIAQAPMPDQDL 161
C+VCGA L D R+D+H G+ H+G K++ + LK+ L + P D+ L
Sbjct 192 CEVCGAFLIVGDAQSRVDDHLMGKQHMGYAKIKATVEELKEKLRKRTEEPDRDERL 247
> 7290737
Length=414
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 104 QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDAL 144
++++ C++CGA L D +R+++H G+ HLG KLR+A+
Sbjct 276 KQMKVCEICGAFLIVGDAQQRIEDHLMGKQHLGYSKLRNAV 316
> ECU10g0840
Length=245
Score = 35.8 bits (81), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 52/115 (45%), Gaps = 25/115 (21%)
Query 52 RLDQAIAQKFKEMEDLGKLGRIDQSMGVSLEL-TSLQTRRAKLLFRNAEG--QLTQRLRP 108
R D A+ K KE E+L R+ +S+G EL S RA F E + +R++
Sbjct 107 RTDPALFGKIKEKEEL--FNRVYESVG---ELGMSGDVERACSAFNECERIREELERIKE 161
Query 109 -------------CDVCGALLSANDTDRRLDEHFSGRVHLGL----QKLRDALKK 146
C +CG L +DT ++D H +GR+H G KL D LKK
Sbjct 162 AYYVKNNGAGMERCGICGVNLVLSDTKAKVDRHLNGRLHRGHLLVRSKLADLLKK 216
> At5g66960
Length=792
Score = 33.9 bits (76), Expect = 0.16, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 104 QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRD 142
Q+ RPC + + + + D D L E F G VH+ ++ +D
Sbjct 250 QKKRPCRIYCSTIGSTDEDVLLHEEFEGNVHVNIRHTKD 288
> Hs5729982
Length=713
Score = 31.6 bits (70), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 43/96 (44%), Gaps = 21/96 (21%)
Query 50 VLRLDQAIAQKFKEMEDLGKLGRIDQSMGVS------LELTSLQTRRAKLLFRNAEGQLT 103
V +L+++ EM+ L QSM S LE+T T LL AE Q+
Sbjct 581 VSKLEESSKATPAEMD----LAHNSQSMHASSASKSVLEVTQKATPNPSLL--AAEDQV- 633
Query 104 QRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQK 139
PC+ CG+L+ D +D HF+ L LQK
Sbjct 634 ----PCEKCGSLVPVWDMPEHMDYHFA----LELQK 661
> At2g37850
Length=447
Score = 31.2 bits (69), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 15/74 (20%)
Query 5 GFEELAVQLCRALVEEADKKVQTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKEM 64
G EE++ Q+ R ++E ++ A+ E KS+ P ++ A+ F+
Sbjct 310 GSEEISSQIQRQFIQE----IELAE-----------ELAKSIEPGNTKMPDAMETIFEAA 354
Query 65 EDLGKLGRIDQSMG 78
DLGKLG + + MG
Sbjct 355 LDLGKLGGVKEVMG 368
> Hs22044051
Length=443
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 97 NAEGQLTQRLRPCDVCGALLSANDTDRRLDEHFSGRVHLGLQKLRDALK-KNLEYIAQAP 155
+ L+Q LR C CG + LD+H+ HLG K + K N EYI++
Sbjct 12 GSSAHLSQGLRSCGGCGTEIKNGQALVALDKHW----HLGCFKCKSCGKLLNAEYISKDG 67
Query 156 MP 157
+P
Sbjct 68 LP 69
> CE20525
Length=1236
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 118 ANDTDRRLDEHFSGRVHLGLQKLRDALKKNLEYI 151
A DT RL +SG+ LG+++ RD L E+I
Sbjct 153 ARDTLYRLSVAYSGQEELGIERARDELDDRREHI 186
> Hs4759312
Length=513
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 35/82 (42%), Gaps = 0/82 (0%)
Query 26 QTAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKEMEDLGKLGRIDQSMGVSLELTS 85
+TA R + SL G + V PEVL + +A ++ G+L + + G L +T
Sbjct 416 ETASTRFEMFSLSGTFGTQYVFPEVLLSENQLAPGEFQVSTDGRLFSLKPTSGPVLTVTL 475
Query 86 LQTRRAKLLFRNAEGQLTQRLR 107
K NA LT + R
Sbjct 476 FGRLYEKDWASNASSGLTAQAR 497
> Hs22044174
Length=1181
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 36/75 (48%), Gaps = 10/75 (13%)
Query 12 QLCRALVEEADKKVQ---TAQMRVDCASLPGVENVKSVHPEVLRLDQAIAQKFKEMEDLG 68
+L R L E A +K+ Q V C G++ VK RL + +++ K+ E +G
Sbjct 697 RLWRLLAEAALQKLDLYTAEQAFVRCKDYQGIKFVK-------RLGKLLSESMKQAEVVG 749
Query 69 KLGRIDQSMGVSLEL 83
GR +++ LE+
Sbjct 750 YFGRFEEAERTYLEM 764
Lambda K H
0.320 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40