bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1589_orf2
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
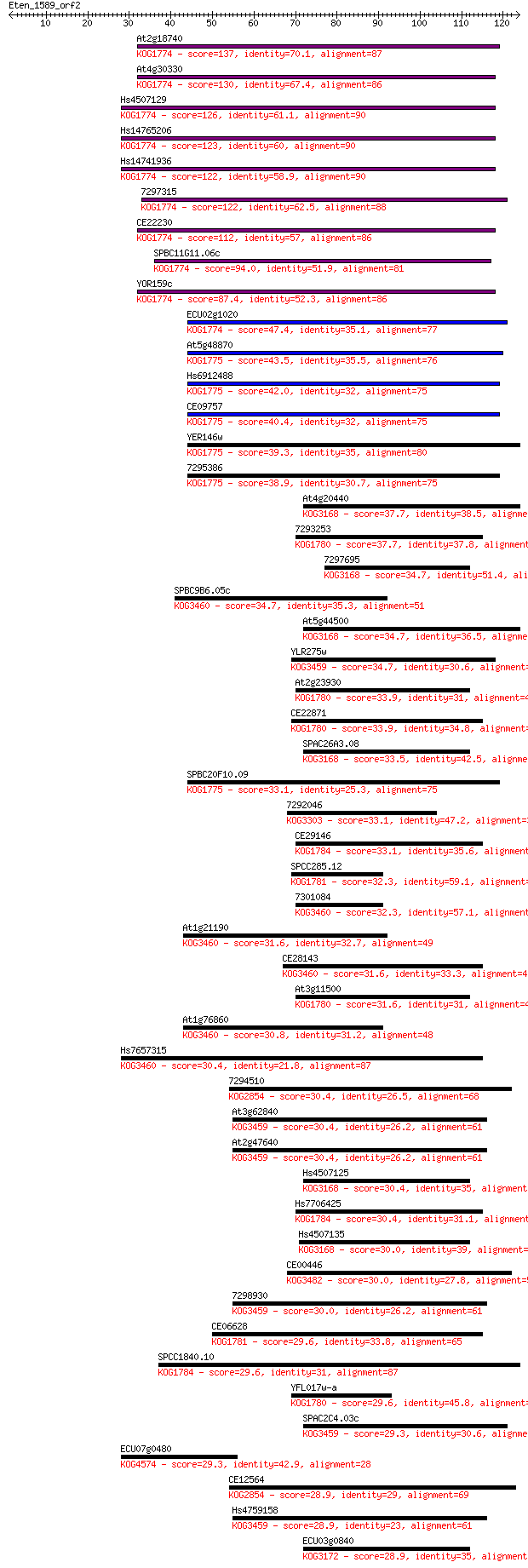
At2g18740 137 5e-33
At4g30330 130 4e-31
Hs4507129 126 8e-30
Hs14765206 123 8e-29
Hs14741936 122 1e-28
7297315 122 1e-28
CE22230 112 1e-25
SPBC11G11.06c 94.0 7e-20
YOR159c 87.4 6e-18
ECU02g1020 47.4 7e-06
At5g48870 43.5 9e-05
Hs6912488 42.0 3e-04
CE09757 40.4 7e-04
YER146w 39.3 0.002
7295386 38.9 0.002
At4g20440 37.7 0.005
7293253 37.7 0.006
7297695 34.7 0.039
SPBC9B6.05c 34.7 0.048
At5g44500 34.7 0.048
YLR275w 34.7 0.048
At2g23930 33.9 0.074
CE22871 33.9 0.079
SPAC26A3.08 33.5 0.093
SPBC20F10.09 33.1 0.12
7292046 33.1 0.14
CE29146 33.1 0.14
SPCC285.12 32.3 0.22
7301084 32.3 0.24
At1g21190 31.6 0.35
CE28143 31.6 0.37
At3g11500 31.6 0.43
At1g76860 30.8 0.56
Hs7657315 30.4 0.77
7294510 30.4 0.87
At3g62840 30.4 0.88
At2g47640 30.4 0.88
Hs4507125 30.4 0.90
Hs7706425 30.4 0.94
Hs4507135 30.0 1.1
CE00446 30.0 1.2
7298930 30.0 1.2
CE06628 29.6 1.5
SPCC1840.10 29.6 1.5
YFL017w-a 29.6 1.6
SPAC2C4.03c 29.3 1.7
ECU07g0480 29.3 1.7
CE12564 28.9 2.2
Hs4759158 28.9 2.2
ECU03g0840 28.9 2.2
> At2g18740
Length=88
Score = 137 bits (345), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 61/87 (70%), Positives = 79/87 (90%), Gaps = 0/87 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
M++ K+Q+IMTQPINLIFRF +K+R+Q+WL+EQ DLRIEGRI GFDEYMN+VLD+AEE+
Sbjct 1 MASTKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEV 60
Query 92 HLKKNTRVFVGRILLKGENISLIMSTG 118
+KKNTR +GRILLKG+NI+L+M+TG
Sbjct 61 SIKKNTRKPLGRILLKGDNITLMMNTG 87
> At4g30330
Length=86
Score = 130 bits (328), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 58/86 (67%), Positives = 76/86 (88%), Gaps = 0/86 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
M++ K+Q+IMTQPINLIFRF +K+R+Q+WL+EQ DLRIEGRI GFDEYMN+VLD+AEE+
Sbjct 1 MASTKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEV 60
Query 92 HLKKNTRVFVGRILLKGENISLIMST 117
+KK TR +GRILLKG+NI+L+M+
Sbjct 61 SIKKKTRKPLGRILLKGDNITLMMNA 86
> Hs4507129
Length=92
Score = 126 bits (317), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 55/90 (61%), Positives = 74/90 (82%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPINLIFR+ N+SR+Q+WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPINLIFRYLQNRSRIQVWLYEQVNMRIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIH K +R +GRI+LKG+NI+L+ S
Sbjct 61 AEEIHSKTKSRKQLGRIMLKGDNITLLQSV 90
> Hs14765206
Length=92
Score = 123 bits (309), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 73/90 (81%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPI+LIFR+ N+SR+Q+WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPISLIFRYLQNRSRIQVWLYEQVNMRIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIH K +R +GRI+LKG NI+L+ S
Sbjct 61 AEEIHSKTKSRKQLGRIMLKGGNITLLQSV 90
> Hs14741936
Length=92
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 53/90 (58%), Positives = 73/90 (81%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M + +K+QK+M QPINLIFR+ N+S++Q+WLYEQ +++IEG I+GFDEYMN+VLDD
Sbjct 1 MAYSGQGQKVQKVMVQPINLIFRYLQNRSQIQVWLYEQVNMQIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIH K +R +GRI+LKG+NI+L+ S
Sbjct 61 AEEIHSKTKSRKQLGRIMLKGDNITLLQSV 90
> 7297315
Length=94
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 70/88 (79%), Gaps = 0/88 (0%)
Query 33 SNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIH 92
N K+QK+M QPINLIFR+ N+SRVQ+WLYE LRIEG I+GFDEYMN+VLDDAEE++
Sbjct 5 GNPKVQKVMVQPINLIFRYLQNRSRVQVWLYENISLRIEGHIVGFDEYMNLVLDDAEEVY 64
Query 93 LKKNTRVFVGRILLKGENISLIMSTGST 120
+K R +GRI+LKG+NI+LI + T
Sbjct 65 VKTRQRRNLGRIMLKGDNITLIQNVSPT 92
> CE22230
Length=90
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 49/86 (56%), Positives = 68/86 (79%), Gaps = 0/86 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MS +KL K+M QP+NLIFR+ N++RVQ+WLYE R+EG I+GFDE+MN+V D+AEE+
Sbjct 1 MSTRKLNKVMVQPVNLIFRYLQNRTRVQIWLYEDVTHRLEGYIIGFDEFMNVVFDEAEEV 60
Query 92 HLKKNTRVFVGRILLKGENISLIMST 117
++K R +GRILLKG+NI+LI +
Sbjct 61 NMKTKGRNKIGRILLKGDNITLIHAA 86
> SPBC11G11.06c
Length=84
Score = 94.0 bits (232), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 61/81 (75%), Gaps = 1/81 (1%)
Query 36 KLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKK 95
++QK+M PIN IF+ + V +WL+EQ D+R++G+I GFDE+MN+VLDDA ++ K
Sbjct 4 RVQKVMIPPINFIFKLLQQHTPVSIWLFEQTDIRLQGQIRGFDEFMNIVLDDAVQVDAKN 63
Query 96 NTRVFVGRILLKGENISLIMS 116
N R +GRILLKG+NI+LI +
Sbjct 64 NKRE-LGRILLKGDNITLIQA 83
> YOR159c
Length=94
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 45/95 (47%), Positives = 63/95 (66%), Gaps = 11/95 (11%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MSNK K M PIN IF F ++ V +WL+EQ +RI+G+I+GFDE+MN+V+D+A EI
Sbjct 1 MSNKVKTKAMVPPINCIFNFLQQQTPVTIWLFEQIGIRIKGKIVGFDEFMNVVIDEAVEI 60
Query 92 ---------HLKKNTRVFVGRILLKGENISLIMST 117
++K T +G+ILLKG+NI+LI S
Sbjct 61 PVNSADGKEDVEKGTP--LGKILLKGDNITLITSA 93
> ECU02g1020
Length=111
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 17/79 (21%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDL--RIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFV 101
P+ I+ F + V++W + D+ + +GRI+GFDEYMN+V+D
Sbjct 48 PLEKIYDFLSRSIDVEIW---EKDMGAKHKGRIVGFDEYMNVVVDGE------------T 92
Query 102 GRILLKGENISLIMSTGST 120
GR LLKG+ I I G+
Sbjct 93 GRYLLKGDCICAIFGKGTC 111
> At5g48870
Length=88
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 6/78 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P LI R +K +W+ + D + G + GFD Y+NMVL+D E + R +
Sbjct 10 PSELIDRCIGSK----IWVIMKGDKELVGILKGFDVYVNMVLEDVTEYEITAEGRRVTKL 65
Query 102 GRILLKGENISLIMSTGS 119
+ILL G NI++++ GS
Sbjct 66 DQILLNGNNIAILVPGGS 83
> Hs6912488
Length=91
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + + D I G ++GFD+++NMVL+D E + R +
Sbjct 14 PLELVDKCI--GSRIHIVM--KSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKL 69
Query 102 GRILLKGENISLIMSTG 118
+ILL G NI++++ G
Sbjct 70 DQILLNGNNITMLVPGG 86
> CE09757
Length=91
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 42/77 (54%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ LI + +K +W+ + D I G + GFD+Y+NMVL+D E + + +
Sbjct 14 PLELIDKCIGSK----IWVIMKNDKEIVGTLTGFDDYVNMVLEDVVEYENTADGKRMTKL 69
Query 102 GRILLKGENISLIMSTG 118
ILL G +I++++ G
Sbjct 70 DTILLNGNHITMLVPGG 86
> YER146w
Length=93
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 49/88 (55%), Gaps = 12/88 (13%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLK-----KNTR 98
P+ +I + K + L Q + EG ++GFD+++N++L+DA E + +N +
Sbjct 8 PLEVIDKTINQKVLIVL----QSNREFEGTLVGFDDFVNVILEDAVEWLIDPEDESRNEK 63
Query 99 VFV--GRILLKGENISLIMSTG-STNTE 123
V GR+LL G NI++++ G T TE
Sbjct 64 VMQHHGRMLLSGNNIAILVPGGKKTPTE 91
> 7295386
Length=91
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 43/77 (55%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + D + G ++GFD+++NM+LDD E + R +
Sbjct 15 PLELVDKCI--GSRIHIIMKN--DKEMVGTLLGFDDFVNMLLDDVTEYENTPDGRRITKL 70
Query 102 GRILLKGENISLIMSTG 118
+ILL G NI++++ G
Sbjct 71 DQILLNGNNITMLVPGG 87
> At4g20440
Length=257
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 31/63 (49%), Gaps = 11/63 (17%)
Query 72 GRIMGFDEYMNMVLDDAEE-----------IHLKKNTRVFVGRILLKGENISLIMSTGST 120
G+ M FD +MN+VL D EE I+ ++ R +G +LL+GE + + G
Sbjct 29 GKFMAFDRHMNLVLGDCEEFRKLPPAKGKKINEEREDRRTLGLVLLRGEEVISMTVEGPP 88
Query 121 NTE 123
E
Sbjct 89 PPE 91
> 7293253
Length=76
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 30/45 (66%), Gaps = 1/45 (2%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENISLI 114
+ G + GFD +MN+VLDD E K NT+ +G ++++G +I ++
Sbjct 27 VTGILRGFDPFMNVVLDDTVE-ECKDNTKNNIGMVVIRGNSIVMV 70
> 7297695
Length=199
Score = 34.7 bits (78), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 9/44 (20%)
Query 77 FDEYMNMVLDDAEEIH--LKKNTRV-------FVGRILLKGENI 111
FD++MN++L D EE KN++V +G +LL+GENI
Sbjct 34 FDKHMNLILGDCEEFRKIRSKNSKVPEREEKRVLGFVLLRGENI 77
> SPBC9B6.05c
Length=93
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 30/51 (58%), Gaps = 4/51 (7%)
Query 41 MTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
+ +P++L+ V+L + D + GR+ +DE++NMVL DAEEI
Sbjct 7 VAEPLDLVRLSLDEIVYVKL----RGDRELNGRLHAYDEHLNMVLGDAEEI 53
> At5g44500
Length=254
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 12/64 (18%)
Query 72 GRIMGFDEYMNMVLDDAEEI------------HLKKNTRVFVGRILLKGENISLIMSTGS 119
G+ M FD +MN+VL D EE + ++ R +G +LL+GE + + G
Sbjct 29 GKFMAFDRHMNLVLGDCEEFRKLPPAKGNKKTNEEREERRTLGLVLLRGEEVISMTVEGP 88
Query 120 TNTE 123
E
Sbjct 89 PPPE 92
> YLR275w
Length=110
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 33/55 (60%), Gaps = 6/55 (10%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIHLKK------NTRVFVGRILLKGENISLIMST 117
+I R+ FD + NMVL++ +E+ +K N F+ ++ L+G+++ +++ T
Sbjct 53 KIIARVKAFDRHCNMVLENVKELWTEKKGKNVINRERFISKLFLRGDSVIVVLKT 107
> At2g23930
Length=80
Score = 33.9 bits (76), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 29/42 (69%), Gaps = 2/42 (4%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
+ G + GFD++MN+V+D+ E++ N + +G ++++G +I
Sbjct 28 VTGTLRGFDQFMNLVVDNTVEVN--GNDKTDIGMVVIRGNSI 67
> CE22871
Length=77
Score = 33.9 bits (76), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENISLI 114
R+ G + GFD +MNMV+D+A E + K V +G +++G ++ ++
Sbjct 26 RVSGILRGFDPFMNMVIDEAVE-YQKDGGSVNLGMTVIRGNSVVIM 70
> SPAC26A3.08
Length=147
Score = 33.5 bits (75), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 10/50 (20%)
Query 72 GRIMGFDEYMNMVLDDAEEI-HLKKNT---------RVFVGRILLKGENI 111
G+++ FD +MN+VL D +E H+KK + +G ++L+GE I
Sbjct 26 GQLLAFDGFMNLVLSDCQEYRHIKKQNVPSNSVYEEKRMLGLVILRGEFI 75
> SPBC20F10.09
Length=80
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 4/75 (5%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGR 103
P+ LI + + LW+ + + G ++GFD+Y+N+VL D E
Sbjct 7 PLELIDKCIGS----NLWVIMKSEREFAGTLVGFDDYVNIVLKDVTEYDTVTGVTEKHSE 62
Query 104 ILLKGENISLIMSTG 118
+LL G + +++ G
Sbjct 63 MLLNGNGMCMLIPGG 77
> 7292046
Length=238
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 23/36 (63%), Gaps = 2/36 (5%)
Query 68 LRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGR 103
L IE R M ++Y LDD E IHLKKN++ ++ R
Sbjct 153 LYIEVRCM--EDYGKFELDDGEVIHLKKNSQHYLPR 186
> CE29146
Length=98
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF---VGRILLKGENISLI 114
I G + GFD+ +N+V++DA E + V +G +++GEN+++I
Sbjct 23 IVGLLKGFDQLINLVIEDAHERSYSETEGVLTTPLGLYIIRGENVAII 70
> SPCC285.12
Length=113
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 69 RIEGRIMGFDEYMNMVLDDAEE 90
+I G + GFD+ MN+VLDD EE
Sbjct 45 QITGILKGFDQLMNLVLDDVEE 66
> 7301084
Length=143
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 70 IEGRIMGFDEYMNMVLDDAEE 90
+ GR+ FD+++NMVL DAEE
Sbjct 7 LRGRLHAFDQHLNMVLGDAEE 27
> At1g21190
Length=97
Score = 31.6 bits (70), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 31/49 (63%), Gaps = 4/49 (8%)
Query 43 QPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
+P++LI + + R+ + L +LR G++ FD+++NM+L D EE+
Sbjct 11 EPLDLIR--LSIEERIYVKLRSDRELR--GKLHAFDQHLNMILGDVEEV 55
> CE28143
Length=102
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 31/60 (51%), Gaps = 12/60 (20%)
Query 67 DLRIEGRIMGFDEYMNMVLDDAEEI------------HLKKNTRVFVGRILLKGENISLI 114
D + GR+ FD+++NMVL + EE + K T+ V + ++G+++ L+
Sbjct 36 DRELRGRLRAFDQHLNMVLSEVEETITTREVDEDTFEEIYKQTKRVVPMLFVRGDSVILV 95
> At3g11500
Length=79
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 28/42 (66%), Gaps = 2/42 (4%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
+ G + GFD++MN+V+D+ E++ T +G ++++G +I
Sbjct 28 VVGTLRGFDQFMNLVVDNTVEVNGDDKTD--IGMVVIRGNSI 67
> At1g76860
Length=98
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 4/48 (8%)
Query 43 QPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE 90
+P++LI + V+L + D + G++ FD+++NM+L D EE
Sbjct 11 EPLDLIRLSLDERIYVKL----RSDRELRGKLHAFDQHLNMILGDVEE 54
> Hs7657315
Length=102
Score = 30.4 bits (67), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 19/99 (19%), Positives = 47/99 (47%), Gaps = 16/99 (16%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M + ++ + +P++LI + V++ + D + GR+ +D+++NM+L D
Sbjct 1 MADDVDQQQTTNTVEEPLDLIRLSLDERIYVKM----RNDRELRGRLHAYDQHLNMILGD 56
Query 88 AEEI------------HLKKNTRVFVGRILLKGENISLI 114
EE + K+T+ + + ++G+ + L+
Sbjct 57 VEETVTTIEIDEETYEEIYKSTKRNIPMLFVRGDGVVLV 95
> 7294510
Length=345
Score = 30.4 bits (67), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query 54 NKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAE----EIH--LKKNTRVFVGRILLK 107
N R+ W+ QP + + +G D Y N++ + A+ ++H +KK+ +L+
Sbjct 69 NSLRIAQWILRQPRVAVFFGCVGEDRYANILKEKAQAAGLDVHYQVKKDVPTGTCAVLIT 128
Query 108 GENISLIMSTGSTN 121
G + SL + + N
Sbjct 129 GTHRSLCANLAAAN 142
> At3g62840
Length=108
Score = 30.4 bits (67), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 38/74 (51%), Gaps = 13/74 (17%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI-------------HLKKNTRVFV 101
K+ Q+ + + + ++ GR+ FD + NMVL++ E+ L N F+
Sbjct 31 KNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVREMWTEVPKTGKGKKKALPVNRDRFI 90
Query 102 GRILLKGENISLIM 115
++ L+G+++ +++
Sbjct 91 SKMFLRGDSVIIVL 104
> At2g47640
Length=108
Score = 30.4 bits (67), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 38/74 (51%), Gaps = 13/74 (17%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI-------------HLKKNTRVFV 101
K+ Q+ + + + ++ GR+ FD + NMVL++ E+ L N F+
Sbjct 31 KNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVREMWTEVPKTGKGKKKALPVNRDRFI 90
Query 102 GRILLKGENISLIM 115
++ L+G+++ +++
Sbjct 91 SKMFLRGDSVIIVL 104
> Hs4507125
Length=231
Score = 30.4 bits (67), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 9/49 (18%)
Query 72 GRIMGFDEYMNMVLDDAEEI---------HLKKNTRVFVGRILLKGENI 111
G FD++MN++L D +E ++ + +G +LL+GEN+
Sbjct 29 GTFKAFDKHMNLILCDCDEFRKIKPKNSKQAEREEKRVLGLVLLRGENL 77
> Hs7706425
Length=96
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 31/48 (64%), Gaps = 3/48 (6%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKN---TRVFVGRILLKGENISLI 114
I G + GFD+ +N++LD++ E + +V +G +++G+N+++I
Sbjct 23 IVGTLKGFDQTINLILDESHERVFSSSQGVEQVVLGLYIVRGDNVAVI 70
> Hs4507135
Length=240
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 14/55 (25%)
Query 71 EGRIM-----GFDEYMNMVLDDAEEI---------HLKKNTRVFVGRILLKGENI 111
+GRI FD++MN++L D +E ++ + +G +LL+GEN+
Sbjct 23 DGRIFIGTFKAFDKHMNLILCDCDEFRKIKPKNAKQPEREEKRVLGLVLLRGENL 77
> CE00446
Length=85
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 68 LRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENISLIMSTGSTN 121
+ +G ++ D YMN+ L AEE ++ N++ +G IL++ N+ + N
Sbjct 29 MEYKGVLVAVDSYMNLQLAHAEE-YIDGNSQGNLGEILIRCNNVLYVGGVDGEN 81
> 7298930
Length=119
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 40/74 (54%), Gaps = 13/74 (17%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI--HLKKNTRV-----------FV 101
K+ Q+ + + + ++ GR+ FD + NMVL++ +E+ L + + F+
Sbjct 36 KNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTELPRTGKGKKKVKPVNKDRFI 95
Query 102 GRILLKGENISLIM 115
++ L+G+++ L++
Sbjct 96 SKMFLRGDSVILVL 109
> CE06628
Length=104
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 11/72 (15%)
Query 50 RFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE-IHLKKNTRVF------VG 102
RF + RV+ Q G + GFD+ +NMVLDD E + +N V +G
Sbjct 18 RFLDKEIRVKF----QGGREASGVLRGFDQLLNMVLDDCREYLRDPQNPSVVGDETRQLG 73
Query 103 RILLKGENISLI 114
I+ +G I+++
Sbjct 74 LIVARGTAITVV 85
> SPCC1840.10
Length=94
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 42/90 (46%), Gaps = 21/90 (23%)
Query 37 LQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE--IHLK 94
L M Q + +I TN RV L G + GFD N++L D+ E I +
Sbjct 3 LADFMEQRVQVI----TNDGRVVL-----------GSLKGFDHTTNLILSDSFERIISMD 47
Query 95 KNTRVF-VGRILLKGENISLIMSTGSTNTE 123
++ +G LL+GEN++++ G N E
Sbjct 48 QDMETIPLGVYLLRGENVAMV---GLVNEE 74
> YFL017w-a
Length=77
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIH 92
++ G + G+D ++N+VLDDA EI+
Sbjct 24 KVAGILRGYDIFLNVVLDDAMEIN 47
> SPAC2C4.03c
Length=115
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/61 (24%), Positives = 30/61 (49%), Gaps = 12/61 (19%)
Query 72 GRIMGFDEYMNMVLDDAEEIHLKK------------NTRVFVGRILLKGENISLIMSTGS 119
R+ FD + NMVL++ +E+ +K N F+ ++ L+G+ + L++ S
Sbjct 55 ARVKAFDRHSNMVLENVKEMWTEKKRTASGKKGKAINKDRFISKMFLRGDGVVLVVRIPS 114
Query 120 T 120
Sbjct 115 A 115
> ECU07g0480
Length=444
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNK 55
+TH++ N +QKI+ +LIF FF +K
Sbjct 373 ITHEIGNYTIQKILRFDPDLIFEFFVSK 400
> CE12564
Length=342
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 34/75 (45%), Gaps = 6/75 (8%)
Query 54 NKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE----IHLKKNTRVFVGRI--LLK 107
N RV W+ P+ + +G D+Y +++ A+E +H + N V G L+
Sbjct 68 NSLRVAQWILNAPNRTVFFGAVGKDQYGDLLASKAKEAGVNVHYQINETVKTGTCAALIN 127
Query 108 GENISLIMSTGSTNT 122
G + SL + NT
Sbjct 128 GTHRSLCAHLAAANT 142
> Hs4759158
Length=118
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/74 (18%), Positives = 38/74 (51%), Gaps = 13/74 (17%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI-------------HLKKNTRVFV 101
K+ Q+ + + + ++ GR+ FD + NMVL++ +E+ N ++
Sbjct 37 KNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKKSKPVNKDRYI 96
Query 102 GRILLKGENISLIM 115
++ L+G+++ +++
Sbjct 97 SKMFLRGDSVIVVL 110
> ECU03g0840
Length=87
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 2/40 (5%)
Query 72 GRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
GR+ D+YMN+VLDD + N+ + +L++G +I
Sbjct 30 GRVREVDDYMNLVLDDV--VVTSNNSSLSRKEVLIRGSSI 67
Lambda K H
0.319 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40