bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1577_orf1
Length=331
Score E
Sequences producing significant alignments: (Bits) Value
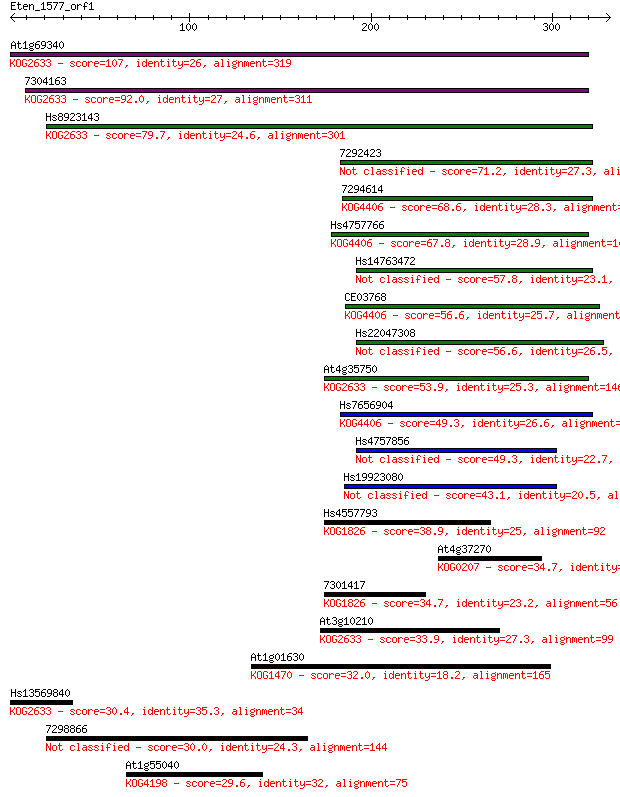
At1g69340 107 2e-23
7304163 92.0 1e-18
Hs8923143 79.7 8e-15
7292423 71.2 3e-12
7294614 68.6 2e-11
Hs4757766 67.8 3e-11
Hs14763472 57.8 3e-08
CE03768 56.6 6e-08
Hs22047308 56.6 8e-08
At4g35750 53.9 5e-07
Hs7656904 49.3 1e-05
Hs4757856 49.3 1e-05
Hs19923080 43.1 7e-04
Hs4557793 38.9 0.014
At4g37270 34.7 0.26
7301417 34.7 0.33
At3g10210 33.9 0.46
At1g01630 32.0 1.8
Hs13569840 30.4 5.8
7298866 30.0 7.7
At1g55040 29.6 8.2
> At1g69340
Length=561
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 83/326 (25%), Positives = 143/326 (43%), Gaps = 14/326 (4%)
Query 1 WMEKMVADVDAVVLVVGCPSDAAIYKQFIPAYFPRDRAEELDASFLLPREVGNANGEIDV 60
++EK + AVV SD IYK+ +P YFPRD EE A LP +VG+ NGE +
Sbjct 221 FLEKQKDKISAVVFCTTTSSDTEIYKRLLPLYFPRDEHEEEVAISKLPADVGDENGETVI 280
Query 61 AERRIRLIAKNESGAGAPAADREAGTTQPVFGGMLARQEKSDDEGDSDSDHDILDSNDVS 120
ER+IR+ A P +P L R+ + + D L +
Sbjct 281 DERKIRIQALPNK---PPPRSFPTPLERPSTDLTLLRRNSNHLDSYLDPAFMSLIKDPDE 337
Query 121 FRVAKAETETEKRLDVLNIKRHGLNH------SSDSSETLYQTYLRQANLLADSAAGAQL 174
R + E + + +K G S+ +L+ YL +AN S +++
Sbjct 338 RRKEQWEKTAQAQSGFNFVKLLGFGDLGGPPLSAAEEYSLHSRYLAKAN----SINLSEI 393
Query 175 QQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTLDPFIRDKYTLLYVNSA 234
+++ +Y G D G P++V + A F +D +L++I+ +P I+ Y+++Y +SA
Sbjct 394 AEMKIVYRGGVDTEGHPVMVVVGAHFLLRCLDLERFVLYVIKEFEPVIQKPYSIVYFHSA 453
Query 235 VHHSNAPSMTLWSEFFGMM-EKFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVY 293
P + ++ K + NL + VLHP KA ++ + VW VY
Sbjct 454 ASLQVQPDLGWMKRLEQILGRKHQRNLQAIYVLHPTFHLKATILTMQFFVDNVVWKKVVY 513
Query 294 LKSIKELSVHAGNHMPKLPNYVVEYD 319
+ +L + +P++V ++D
Sbjct 514 ADRLLQLFKYVPREQLTIPDFVFQHD 539
> 7304163
Length=482
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 84/342 (24%), Positives = 141/342 (41%), Gaps = 36/342 (10%)
Query 9 VDAVVLVVGCPSDAAIYKQFIPAYFPRDRAEELDASFLLPREVGNANGEIDVAE--RRIR 66
+ V+L VG S+ Y+ P YFPRD+ EE A + LP+++G GE + R+IR
Sbjct 137 LQVVILCVGS-SERGTYEVLAPLYFPRDQLEERSALWQLPKDIGGEFGEPQHPDPDRQIR 195
Query 67 LIAK-------------------------NESGAGAPAADREAGTTQPVFGGMLARQEKS 101
+I N S A D + G+ A+ ++
Sbjct 196 IIRNPQHSVHMRHHLADDDSDVSPHDMEGNSSDLEYGARDMNGLSLNSYSSGLQAQLQRD 255
Query 102 DDEGD--SDSDHDILDSNDVSFRVAKAETETEKRLDVLNIKRHGL-NHSSDSSETLYQTY 158
D SD + N +S V E + + L + +S E Y+
Sbjct 256 LDRQHLLSDRPRTGVYENVISEGVEGIEHQESPMQETTGDMNDVLPDELMESPEERYERL 315
Query 159 LRQANLLADSAAGAQLQQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTL 218
LR+A + ++ + +Y SG D GRP+IVF FPA +D LL++I+ L
Sbjct 316 LRRAQV----EDLTEVSGIGCLYQSGVDRLGRPVIVFCGKWFPAQNIDLEKALLYLIKLL 371
Query 219 DPFIRDKYTLLYVNSAVHHSNAPSMTLWSEFFGMME-KFEDNLDQLLVLHPGLLFKAAFT 277
DP ++ Y + Y ++ +N PS+ E + ++ K++ NL ++HP K
Sbjct 372 DPIVKGDYVISYFHTLTSTNNYPSLHWLREVYSVLPYKYKKNLKAFYIVHPTFWTKMMTW 431
Query 278 CCWPYLASHVWNGTVYLKSIKELSVHAGNHMPKLPNYVVEYD 319
++A + L ++ L ++P Y+ EYD
Sbjct 432 WFTTFMAPAIKAKVHSLPGVEHLYSAITKDQLEIPAYITEYD 473
> Hs8923143
Length=497
Score = 79.7 bits (195), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 74/305 (24%), Positives = 128/305 (41%), Gaps = 37/305 (12%)
Query 21 DAAIYKQFIPAYFPRDRAEELDASFLLPREVGNANGEIDVAERRIRLIAKNESGAGAPAA 80
+ Y++ +P YFPR EE + LP ++GNA GE V ER+IR+ K GAP
Sbjct 211 EEGTYQKLLPLYFPRSLKEENRSLPYLPADIGNAEGEPVVPERQIRISEK----PGAPED 266
Query 81 DREAGTTQPVFGGMLARQEKSDDEGDSDSDHDILDSNDVSFRVAKAETETEKRLDVLNIK 140
++E +DEG D+SF + A E +D +
Sbjct 267 NQE-----------------EEDEG---------LGVDLSFIGSHAFARMEGDIDK---Q 297
Query 141 RHGLNHSSDSSETLYQTYLRQAN---LLADSAAGAQLQQLQFIYPSGHDNAGRPIIVFLA 197
R + S L + + R N A S + + L+ +Y +G DN GR ++V +
Sbjct 298 RKLILQGQLSEAALQKQHQRNYNRWLCQARSEDLSDIASLKALYQTGVDNCGRTVMVVVG 357
Query 198 AVFPAATVDSYILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAPSMTLWSEFFGMME-KF 256
P +D LL+ I +D +Y L+Y ++ N + + +++ K+
Sbjct 358 RNIPVTLIDMDKALLYFIHVMDHIAVKEYVLVYFHTLTSEYNHLDSDFLKKLYDVVDVKY 417
Query 257 EDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELSVHAGNHMPKLPNYVV 316
+ NL + +HP K + + S + + ++ S+ +L P +V+
Sbjct 418 KRNLKAVYFVHPTFRSKVSTWFFTTFSVSGLKDKIHHVDSLHQLFSAISPEQIDFPPFVL 477
Query 317 EYDAK 321
EYDA+
Sbjct 478 EYDAR 482
> 7292423
Length=468
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/145 (26%), Positives = 71/145 (48%), Gaps = 9/145 (6%)
Query 183 SGHDNAGRPIIVFLAAVFPAATVDSYI-----LLLHIIRTLDPFIRDKYTLLYVNSAVHH 237
SG NA I++F A P + Y L L++++TL+ + D Y L+Y++ +
Sbjct 325 SGGQNA---IVIFCACHLPDRSRADYSYVMDNLFLYVVKTLEQLVTDDYVLIYLHGGSNR 381
Query 238 SNAPSMTLWSEFFGMMEK-FEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKS 296
N P + ++++ +L + ++HP K+ P++++ W VY+KS
Sbjct 382 RNVPPFPWLKRCYQLLDRRLRKSLKHMYLVHPTFWIKSLVWMARPFVSTKFWRKLVYVKS 441
Query 297 IKELSVHAGNHMPKLPNYVVEYDAK 321
++EL +H +P V +YDAK
Sbjct 442 LEELGMHVVVEKAAIPEKVKQYDAK 466
> 7294614
Length=476
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 68/140 (48%), Gaps = 4/140 (2%)
Query 184 GHDNAGRPIIVFLAAVFPAAT-VDSYILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAPS 242
G D GR I A+ FP + ++ ++ II+ ++PF+ + Y L+Y + + N PS
Sbjct 105 GTDKQGRHIFGIYASRFPEKSQLEGFVR--EIIKEIEPFVENDYILVYFHQGLKEDNKPS 162
Query 243 MT-LWSEFFGMMEKFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELS 301
LW+ + + F NL L V+HP + + P+++ VY+ S+ EL
Sbjct 163 AQFLWNSYKELDRNFRKNLKTLYVVHPTWFIRVIWNFFSPFISDKFRKKLVYISSLDELR 222
Query 302 VHAGNHMPKLPNYVVEYDAK 321
G + KLP+ + + D K
Sbjct 223 QALGLNKLKLPDNICDLDDK 242
> Hs4757766
Length=439
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 41/144 (28%), Positives = 70/144 (48%), Gaps = 2/144 (1%)
Query 178 QFIYPSGHDNAGRPIIVFLAAVFPAA-TVDSYILLLHIIRTLDPFIRDKYTLLYVNSAVH 236
Q + +G D GR IIVF A P + +D LL ++ TLD ++ YTLLY++ +
Sbjct 71 QIVEVAGDDKYGRKIIVFSACRMPPSHQLDHSKLLGYLKHTLDQYVESDYTLLYLHHGLT 130
Query 237 HSNAPSMTLWSEFFGMME-KFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLK 295
N PS++ + + + K++ N+ L ++HP + K P ++ Y+
Sbjct 131 SDNKPSLSWLRDAYREFDRKYKKNIKALYIVHPTMFIKTLLILFKPLISFKFGQKIFYVN 190
Query 296 SIKELSVHAGNHMPKLPNYVVEYD 319
+ ELS H +P V++YD
Sbjct 191 YLSELSEHVKLEQLGIPRQVLKYD 214
> Hs14763472
Length=371
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/136 (22%), Positives = 66/136 (48%), Gaps = 6/136 (4%)
Query 192 IIVFLAAVFPAATVDSY-----ILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAPSMTLW 246
IIVF A P +++ Y L L++I +L+ + + Y ++Y+N A P +
Sbjct 191 IIVFAACFLPDSSLPDYHYIMENLFLYVISSLELLVAEDYMIVYLNGATPRRRMPGIGWL 250
Query 247 SEFFGMMEK-FEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELSVHAG 305
+ + M+++ NL L+++HP + P+++ N Y+ S+++L
Sbjct 251 KKCYQMIDRRLRKNLKSLIIVHPSWFIRTVLAISRPFISVKFINKIQYVHSLEDLEQLIP 310
Query 306 NHMPKLPNYVVEYDAK 321
++P+ V++Y+ +
Sbjct 311 MEHVQIPDCVLQYEEE 326
> CE03768
Length=444
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 68/142 (47%), Gaps = 2/142 (1%)
Query 186 DNAGRPIIVFLAAVFPAAT-VDSYILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAPSMT 244
D GRPI+V A P++ +D LL ++++ +D + YT++Y + + N P +
Sbjct 89 DRVGRPIVVVYAYRLPSSKEIDHARLLQYLVQIIDKIVDQDYTIVYFHYGLRSHNKPPVR 148
Query 245 -LWSEFFGMMEKFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELSVH 303
L+ + + +F+ NL L V+HP + F+ +++S N Y+ I EL
Sbjct 149 WLFQAYKQLDRRFKKNLKALYVVHPTRFIRIIFSLFKGFISSKFENKFHYVMCIDELENA 208
Query 304 AGNHMPKLPNYVVEYDAKPKTK 325
LP+ + ++D T+
Sbjct 209 LSVARLNLPSPIRDHDKSFSTQ 230
> Hs22047308
Length=771
Score = 56.6 bits (135), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 65/149 (43%), Gaps = 13/149 (8%)
Query 192 IIVFLAAVFPAATVDSY-----ILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAPSMTLW 246
IIVF A P ++ Y L L++I TL+ + + Y ++Y+N A P +
Sbjct 602 IIVFAACFLPDSSRADYHYVMENLFLYVISTLELMVAEDYMIVYLNGATPRRRMPGLGWM 661
Query 247 SEFFGMME-KFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELS---- 301
+ + M++ + NL +++HP + P+++S + Y+ S+ ELS
Sbjct 662 KKCYQMIDRRLRKNLKSFIIVHPSWFIRTILAVTRPFISSKFSSKIKYVNSLSELSGLIP 721
Query 302 ---VHAGNHMPKLPNYVVEYDAKPKTKIL 327
+H + KL + E KT L
Sbjct 722 MDCIHIPESIIKLDEELREASEAAKTSCL 750
> At4g35750
Length=202
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/150 (24%), Positives = 70/150 (46%), Gaps = 4/150 (2%)
Query 174 LQQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTLDPFI-RDKYTLLYVN 232
+++L+ G D GR I+ + FPA + +L ++ + P + R + +LYV+
Sbjct 14 IEKLEIFKIHGRDKRGRKILRIIGKFFPARFLSLDVLKKYLEEKIFPRLGRKPFAVLYVH 73
Query 233 SAVHHS-NAPSMTLWSEFFGMME-KFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNG 290
+ V S N P ++ + + DNL ++ LHPGL + C +L S G
Sbjct 74 TGVQRSENFPGISALRAIYDAIPVNVRDNLQEVYFLHPGLQSRLFLATCGRFLFSGGLYG 133
Query 291 TV-YLKSIKELSVHAGNHMPKLPNYVVEYD 319
+ Y+ + L H + ++P +V ++D
Sbjct 134 KLRYISRVDYLWEHVRRNEIEMPEFVYDHD 163
> Hs7656904
Length=464
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/172 (21%), Positives = 66/172 (38%), Gaps = 33/172 (19%)
Query 183 SGHDNAGRPIIVFLAAVFPAA-TVDSYILLLHIIRTLDPFIRDKYTLLYVNSAVHHSNAP 241
+G D GR ++ F P + +D LL ++ TLD ++ + YT++Y + ++ N P
Sbjct 26 AGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYLKYTLDQYVENDYTIVYFHYGLNSRNKP 85
Query 242 S-----------------MTLWSEFFGMME---------------KFEDNLDQLLVLHPG 269
S +T+W + +++ NL L V+HP
Sbjct 86 SLGWLQSAYKEFDRKDGDLTMWPRLVSNSKLKRSSHLSLPKYWDYRYKKNLKALYVVHPT 145
Query 270 LLFKAAFTCCWPYLASHVWNGTVYLKSIKELSVHAGNHMPKLPNYVVEYDAK 321
K + P ++ +Y + EL H +P V+ YD K
Sbjct 146 SFIKVLWNILKPLISHKFGKKVIYFNYLSELHEHLKYDQLVIPPEVLRYDEK 197
> Hs4757856
Length=314
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/116 (21%), Positives = 55/116 (47%), Gaps = 6/116 (5%)
Query 192 IIVFLAAVFPAATVDSYILLL-----HIIRTLDPFIRDKYTLLYVNSAVHHSNAPSMTLW 246
I+VF P ++ +Y L+ ++I TL+ + + Y ++Y+N A PS+
Sbjct 167 IVVFAVCFMPESSQPNYRYLMDNLFKYVIGTLELLVAENYMIVYLNGATTRRKMPSLGWL 226
Query 247 SEFFGMME-KFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIKELS 301
+ + ++ + NL L+++HP + P+++S Y+ ++ EL+
Sbjct 227 RKCYQQIDRRLRKNLKSLIIVHPSWFIRTLLAVTRPFISSKFSQKIRYVFNLAELA 282
> Hs19923080
Length=275
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/123 (19%), Positives = 58/123 (47%), Gaps = 6/123 (4%)
Query 185 HDNAGRPIIVFLAAVFPAATVDSYI-----LLLHIIRTLDPFIRDKYTLLYVNSAVHHSN 239
H + +I+F + P +++ +Y L +++ TL+ + + Y L++++ +
Sbjct 126 HGDGLNAVILFASCYLPRSSIPNYTYVMEHLFRYMVGTLELLVAENYLLVHLSGGTSRAQ 185
Query 240 APSMTLWSEFFGMMEK-FEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIK 298
P ++ + + +++ NL L+V+H KA P+++S +L S+
Sbjct 186 VPPLSWIRQCYRTLDRRLRKNLRALVVVHATWYVKAFLALLRPFISSKFTRKIRFLDSLG 245
Query 299 ELS 301
EL+
Sbjct 246 ELA 248
> Hs4557793
Length=2818
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 1/93 (1%)
Query 174 LQQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTLDPFIRDKYTLLYVNS 233
L+ L Y +G AG PI ++A F ++ +L+ H++ TL P+ Y ++ +
Sbjct 1566 LKTLSIFYQAGTSKAGNPIFYYVARRFKTGQINGDLLIYHVLLTLKPYYAKPYEIVVDLT 1625
Query 234 AVHHSNAPSMTLWSEFFGMMEKFE-DNLDQLLV 265
SN S++F + F DN+ + +
Sbjct 1626 HTGPSNRFKTDFLSKWFVVFPGFAYDNVSAVYI 1658
> At4g37270
Length=819
Score = 34.7 bits (78), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 28/57 (49%), Gaps = 5/57 (8%)
Query 237 HSNAPSMTLWSEFFGMMEKFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVY 293
HSN P + W + FG E+ ++VL + F F WP+L++ G+VY
Sbjct 349 HSNKPKLQRWLDEFG-----ENYSKVVVVLSLAIAFLGPFLFKWPFLSTAACRGSVY 400
> 7301417
Length=2802
Score = 34.7 bits (78), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 13/56 (23%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 174 LQQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTLDPFIRDKYTLL 229
L+ + Y +G +G P+ ++A + + +L+ H+I TL PF + ++
Sbjct 1618 LKSMNIFYQAGTSKSGYPVFYYIARRYKIGETNGDLLIYHVILTLKPFCHSPFEVV 1673
> At3g10210
Length=237
Score = 33.9 bits (76), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 46/102 (45%), Gaps = 3/102 (2%)
Query 172 AQLQQLQFIYPSGHDNAGRPIIVFLAAVFPAATVDSYILLLHIIRTL-DPFIRDKYTLLY 230
+ L LQF G D +G I + FPA V + L +I + + + L+Y
Sbjct 47 SDLDLLQFFTLQGLDRSGNRIFRIVGKYFPARVVSAERLKKYISQKISNQCPEGPLCLVY 106
Query 231 VNSAVHH-SNAPSMTLWSEFF-GMMEKFEDNLDQLLVLHPGL 270
++S V N+P +T+ + + +D L + +HPGL
Sbjct 107 MHSTVQKDDNSPGITILRWIYEDLPSDIKDRLQLVYFIHPGL 148
> At1g01630
Length=255
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 30/171 (17%), Positives = 69/171 (40%), Gaps = 13/171 (7%)
Query 134 LDVLNIKRHGLNHSSDSSETLYQTYLRQANLLADSAAGAQLQQLQFIYPSGHDNAGRPII 193
LD+ LN+ + L + ++ +A + D + Q GHD GRPI
Sbjct 62 LDIEKASTMFLNYLTWKRSMLPKGHIPEAEIANDLSHNKMCMQ-------GHDKMGRPIA 114
Query 194 VFLAAVFPAATVDSYILLLHIIRTLD------PFIRDKYTLLYVNSAVHHSNAPSMTLWS 247
V + + + ++ TL+ P ++K+ + +SN +
Sbjct 115 VAIGNRHNPSKGNPDEFKRFVVYTLEKICARMPRGQEKFVAIGDLQGWGYSNCDIRGYLA 174
Query 248 EFFGMMEKFEDNLDQLLVLHPGLLFKAAFTCCWPYLASHVWNGTVYLKSIK 298
+ + + + L +L ++H +F A+ +P++ ++ V++++ K
Sbjct 175 ALSTLQDCYPERLGKLYIVHAPYIFMTAWKVIYPFIDANTKKKIVFVENKK 225
> Hs13569840
Length=325
Score = 30.4 bits (67), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 1 WMEKMVADVDAVVLVVGCPSDAAIYKQFIPAYFP 34
W+E+ VD +++ V D IY+ +P YFP
Sbjct 290 WLEQHKDKVDRLIICVFLEKDEDIYRSRLPHYFP 323
> 7298866
Length=782
Score = 30.0 bits (66), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 35/148 (23%), Positives = 64/148 (43%), Gaps = 13/148 (8%)
Query 21 DAAIYKQFIPAYFPRDRAEELDASFLLPREVGNANGEIDVAERRIRLIAKNESGAGAPAA 80
D +Y+Q+I + A++ + S +E+ + G++D + R + + ++G GA
Sbjct 386 DGGVYQQYI---VEKTLAQKGNVS---TQELED-EGKVDKRDERRKKASSGKAGGGAQGR 438
Query 81 DREAGTTQPVFGGMLARQEKSDDEGDSDSDHDILDSNDVSFRVAK----AETETEKRLDV 136
+ + +T+ G A Q SDDE D+ S + K +T +L
Sbjct 439 ETKTKSTKKHQRGKAAAQFDSDDEDDAQQGSR--GGGGASKKAVKPLELVKTADIVKLIT 496
Query 137 LNIKRHGLNHSSDSSETLYQTYLRQANL 164
+++ GL H S S +LY Q L
Sbjct 497 ASLEEEGLEHLSKSIASLYTNQFNQTAL 524
> At1g55040
Length=849
Score = 29.6 bits (65), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 10/79 (12%)
Query 65 IRLIAKNE----SGAGAPAADREAGTTQPVFGGMLARQEKSDDEGDSDSDHDILDSNDVS 120
+R +++N+ G G PA DR+ VF G L R+ DEGD S + +S + +
Sbjct 221 LRSLSRNDIQILVGHGCPATDRKV-----VFSGKLLRKRVHLDEGDVCSSCSLRNSCEKA 275
Query 121 FRVAKAETETEKRLDVLNI 139
F + E E + +D++ I
Sbjct 276 FLLTNKEDEA-RTIDLMRI 293
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7812669732
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40