bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1569_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
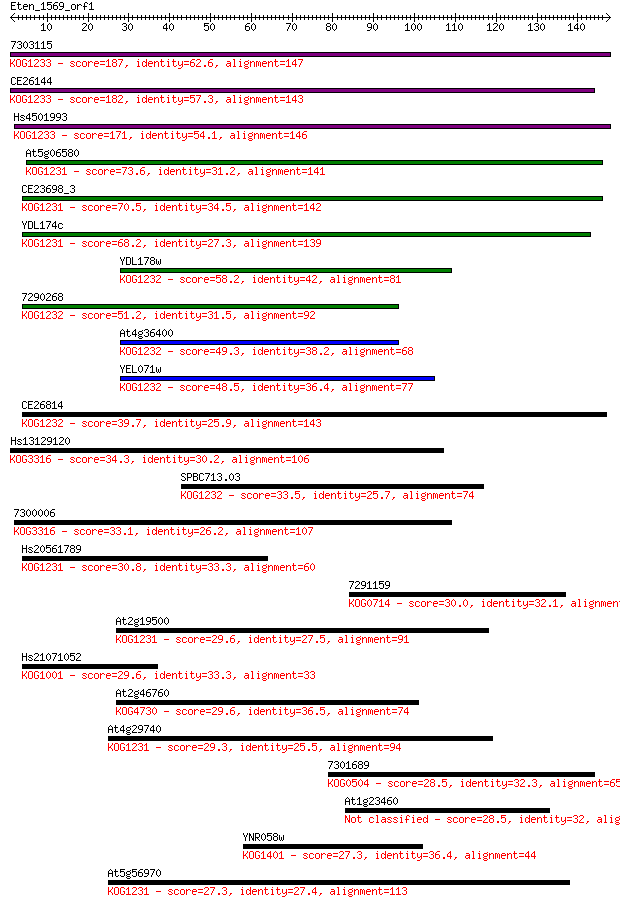
7303115 187 5e-48
CE26144 182 2e-46
Hs4501993 171 4e-43
At5g06580 73.6 1e-13
CE23698_3 70.5 1e-12
YDL174c 68.2 6e-12
YDL178w 58.2 6e-09
7290268 51.2 6e-07
At4g36400 49.3 3e-06
YEL071w 48.5 5e-06
CE26814 39.7 0.002
Hs13129120 34.3 0.083
SPBC713.03 33.5 0.13
7300006 33.1 0.20
Hs20561789 30.8 1.1
7291159 30.0 1.7
At2g19500 29.6 1.9
Hs21071052 29.6 2.2
At2g46760 29.6 2.2
At4g29740 29.3 2.4
7301689 28.5 4.4
At1g23460 28.5 4.5
YNR058w 27.3 9.6
At5g56970 27.3 9.9
> 7303115
Length=631
Score = 187 bits (476), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 92/147 (62%), Positives = 109/147 (74%), Gaps = 0/147 (0%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSK 60
VG LE LR G+T+GHEPDS EFST+GGWVATRASGMKKN YGNIED+VV V +VT
Sbjct 236 VGQDLERVLRSEGLTVGHEPDSYEFSTLGGWVATRASGMKKNVYGNIEDLVVRVRMVTPS 295
Query 61 GSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
G+L ++ SAPRVS GP ++LGSEGTLG+IT+V+LKV+ LP + YG LAFP F GV
Sbjct 296 GTLERECSAPRVSCGPDFNHVILGSEGTLGVITEVVLKVRPLPSLRRYGSLAFPNFEQGV 355
Query 121 HFLRHVALDNLQPASIRLMDKTQTQCG 147
F+R VA QPAS+RLMD Q G
Sbjct 356 LFMREVARRRCQPASVRLMDNEQFMFG 382
> CE26144
Length=597
Score = 182 bits (461), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 82/143 (57%), Positives = 108/143 (75%), Gaps = 0/143 (0%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSK 60
VG SLE +L + G T GHEPDS+EFST+GGWV+TRASGMKKN+YGNIED+VV + V K
Sbjct 212 VGQSLERQLNKKGFTCGHEPDSIEFSTLGGWVSTRASGMKKNKYGNIEDLVVHLNFVCPK 271
Query 61 GSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
G + +Q PR+SSGP + Q++LGSEGTLG++++V +K+ +PE K +G FP F +GV
Sbjct 272 GIIQKQCQVPRMSSGPDIHQIILGSEGTLGVVSEVTIKIFPIPEVKRFGSFVFPNFESGV 331
Query 121 HFLRHVALDNLQPASIRLMDKTQ 143
+F R VA+ QPAS+RLMD Q
Sbjct 332 NFFREVAIQRCQPASLRLMDNDQ 354
> Hs4501993
Length=658
Score = 171 bits (433), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 79/146 (54%), Positives = 103/146 (70%), Gaps = 0/146 (0%)
Query 2 GASLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKG 61
G LE +L+ G GHEPDS+EFSTVGGWV+TRASGMKKN YGNIED+VV + +VT +G
Sbjct 284 GQELERQLKESGYCTGHEPDSLEFSTVGGWVSTRASGMKKNIYGNIEDLVVHIKMVTPRG 343
Query 62 SLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVH 121
+ + PR+S+GP + ++GSEGTLG+IT+ +K++ +PE + YG +AFP F GV
Sbjct 344 IIEKSCQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPVPEYQKYGSVAFPNFEQGVA 403
Query 122 FLRHVALDNLQPASIRLMDKTQTQCG 147
LR +A PASIRLMD Q Q G
Sbjct 404 CLREIAKQRCAPASIRLMDNKQFQFG 429
> At5g06580
Length=417
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 74/142 (52%), Gaps = 3/142 (2%)
Query 5 LEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLN 64
L E L +G+ +P +++GG ATR SG RYG + D V+ + VV G +
Sbjct 73 LNEYLEEYGLFFPLDPGP--GASIGGMCATRCSGSLAVRYGTMRDNVISLKVVLPNGDVV 130
Query 65 QQQSAPRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFL 123
+ S R S+ G L +L++GSEGTLG+IT++ L+++ +P+ FPT
Sbjct 131 KTASRARKSAAGYDLTRLIIGSEGTLGVITEITLRLQKIPQHSVVAVCNFPTVKDAADVA 190
Query 124 RHVALDNLQPASIRLMDKTQTQ 145
+ +Q + + L+D+ Q +
Sbjct 191 IATMMSGIQVSRVELLDEVQIR 212
> CE23698_3
Length=454
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 78/146 (53%), Gaps = 6/146 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
+L + ++ G+ +P + ++V G VAT ASG RYG +++ VV++ VV + G++
Sbjct 108 ALNDAIKNSGLFFPVDPGAD--ASVCGMVATSASGTNAIRYGTMKENVVNLEVVLADGTI 165
Query 64 NQQQS---APRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAG 119
+ PR SS G + +L +GSEGTLGIIT+ +KV P+ + +FPT
Sbjct 166 IDTKGKGRCPRKSSAGFNFTELFVGSEGTLGIITEATVKVHPRPQFLSAAVCSFPTVHEA 225
Query 120 VHFLRHVALDNLQPASIRLMDKTQTQ 145
+ V N+ A I L+D Q Q
Sbjct 226 ASTVVEVLQWNIPVARIELLDTVQIQ 251
> YDL174c
Length=587
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/140 (27%), Positives = 73/140 (52%), Gaps = 3/140 (2%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
L + L HG+ G +P + +GG +A SG RYG +++ ++++ +V G++
Sbjct 230 DLNDYLSDHGLMFGCDPGPG--AQIGGCIANSCSGTNAYRYGTMKENIINMTIVLPDGTI 287
Query 64 NQQQSAPRVSS-GPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHF 122
+ + PR SS G +L L +GSEGTLGI+T+ +K + P+ + ++F T
Sbjct 288 VKTKKRPRKSSAGYNLNGLFVGSEGTLGIVTEATVKCHVKPKAETVAVVSFDTIKDAAAC 347
Query 123 LRHVALDNLQPASIRLMDKT 142
++ + ++ L+D+
Sbjct 348 ASNLTQSGIHLNAMELLDEN 367
> YDL178w
Length=530
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 34/82 (41%), Positives = 51/82 (62%), Gaps = 3/82 (3%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL-NQQQSAPRVSSGPSLQQLMLGSE 86
VGG VAT A G++ RYG++ V+ + VV G + N S + ++G L+QL +GSE
Sbjct 202 VGGVVATNAGGLRLLRYGSLHGSVLGLEVVMPNGQIVNSMHSMRKDNTGYDLKQLFIGSE 261
Query 87 GTLGIITQVLLKVKLLPECKAY 108
GT+GIIT V + +P+ KA+
Sbjct 262 GTIGIITGV--SILTVPKPKAF 281
> 7290268
Length=533
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS- 62
+ ++R R G+T+ + + +GG V+T A G++ RYGN+ V+ V V + G
Sbjct 190 NFDQRAREVGLTVPLDLGAKASCHIGGNVSTNAGGVRVVRYGNLHGSVLGVEAVLATGQV 249
Query 63 LNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQV 95
L+ + + ++G ++ L +GSEGTLG++T++
Sbjct 250 LDLMSNFKKDNTGYHMKHLFIGSEGTLGVVTKL 282
> At4g36400
Length=524
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSE 86
+GG V+T A G++ RYG++ V+ + VT+ G+ L+ + + ++G L+ L +GSE
Sbjct 202 IGGNVSTNAGGLRLIRYGSLHGTVLGLEAVTANGNVLDMLGTLRKDNTGYDLKHLFIGSE 261
Query 87 GTLGIITQV 95
G+LGI+T+V
Sbjct 262 GSLGIVTKV 270
> YEL071w
Length=496
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 46/78 (58%), Gaps = 1/78 (1%)
Query 28 VGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPRV-SSGPSLQQLMLGSE 86
VGG V+T A G+ RYG++ V+ + VV G + +A R ++G L+QL +G+E
Sbjct 168 VGGVVSTNAGGLNFLRYGSLHGNVLGLEVVLPNGEIISNINALRKDNTGYDLKQLFIGAE 227
Query 87 GTLGIITQVLLKVKLLPE 104
GT+G++T V + P+
Sbjct 228 GTIGVVTGVSIVAAAKPK 245
> CE26814
Length=487
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 66/148 (44%), Gaps = 6/148 (4%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGS- 62
L+ +L + G + + + +GG +AT A G++ RYG++ ++ + VV
Sbjct 143 DLDNKLAKLGYMMPFDLGAKGSCQIGGNIATCAGGIRLIRYGSLHAHLLGLTVVLPDEHG 202
Query 63 --LNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGV 120
L+ S + ++ L LGSEG LG+IT V + P+ L +F
Sbjct 203 TVLHLGSSIRKDNTTLHTPHLFLGSEGQLGVITSVTMTAVPKPKSVQSAMLGIESFKKCC 262
Query 121 HFLRHVALDNLQP--ASIRLMDKTQTQC 146
L+ +A +L +S L+D +C
Sbjct 263 EVLK-LAKSSLTEILSSFELLDDATMEC 289
> Hs13129120
Length=173
Score = 34.3 bits (77), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 8/111 (7%)
Query 1 VGASLEERLRRHGVTLGHEPDSMEFSTVGGWVAT---RASGMKKNRYGN--IEDMVVDVV 55
VG +L ERL R + E D ++F WVA + ++ N G ++D ++
Sbjct 58 VGQALGERLPRETLAFREELDVLKFLCKDLWVAVFQKQMDSLRTNHQGTYVLQDNSFPLL 117
Query 56 VVTSKGSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECK 106
+ + G L + AP+ + L+ G+ TLGI + V V LP CK
Sbjct 118 LPMASG-LQYLEEAPKFLAFTC--GLLRGALYTLGIESVVTASVAALPVCK 165
> SPBC713.03
Length=526
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Query 43 RYGNIEDMVVDVVVVTSKGS-LNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKL 101
RYG++ ++ + V G+ L+ + + ++G ++QL +GSEG LG+IT++ +
Sbjct 212 RYGSLHGSILGMEAVLPDGTILDNLVTLRKDNTGLDIKQLFIGSEGYLGVITKLSVICPK 271
Query 102 LPECKAYGCLAFPTF 116
P P++
Sbjct 272 RPSSTNVAFFGVPSY 286
> 7300006
Length=152
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 50/112 (44%), Gaps = 8/112 (7%)
Query 2 GASLEERLRRHGVTLGHEPDSMEFSTVGGWV---ATRASGMKKNRYGN--IEDMVVDVVV 56
G L ERL R E ++M+F W+ + ++ N +G ++D +
Sbjct 38 GYRLIERLTREVSRFKDELETMKFICTDFWMLIYKKQVDNLRTNNHGMYVVQDKAFRFLT 97
Query 57 VTSKGSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKLLPECKAY 108
S G+ Q + AP+ + L+ G+ LGI + V +V+ +P CK +
Sbjct 98 RISPGT-KQLEHAPKFVAFTC--GLVRGALSNLGINSTVTAEVQSIPACKFH 146
> Hs20561789
Length=230
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL 63
+L LR G+ +P + +++ G AT ASG RYG + D V+++ VV G L
Sbjct 162 ALNAHLRDSGLWFPVDPGA--DASLCGMAATGASGTNAVRYGTMRDNVLNLEVVLPDGRL 219
> 7291159
Length=342
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 8/53 (15%)
Query 84 GSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFLRHVALDNLQPASI 136
G G G+ +L VK+ P CKA + FP + L NL+PA++
Sbjct 191 GRNGVDGVQYDRILTVKIPPGCKAGTKICFPN--------EGIQLPNLEPANV 235
> At2g19500
Length=515
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 44/93 (47%), Gaps = 8/93 (8%)
Query 27 TVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSL--NQQQSAPRVSSGPSLQQLMLG 84
TVGG ++ G + R G + V+++ V+T KG + +Q P + G +LG
Sbjct 155 TVGGTLSNGGIGGQVFRNGPLVSNVLELDVITGKGEMLTCSRQLNPELFYG------VLG 208
Query 85 SEGTLGIITQVLLKVKLLPECKAYGCLAFPTFT 117
G GIIT+ + + P+ + + + FT
Sbjct 209 GLGQFGIITRARIVLDHAPKRAKWFRMLYSDFT 241
> Hs21071052
Length=1009
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 4 SLEERLRRHGVTLGHEPDSMEFSTVGGWVATRA 36
++ ++L++HG LG P ++ F+ GW + RA
Sbjct 159 AVSDQLKKHGFKLGPAPKTLGFNLESGWGSGRA 191
> At2g46760
Length=603
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 5/78 (6%)
Query 27 TVGGWVATRASGMKKNRYGN-IEDMVVDVVVVTSKGSLNQQQSAPRV---SSGPSLQQLM 82
TVGG + T A G G+ + D V ++ +V S GS+N + RV ++ P
Sbjct 168 TVGGMMGTGAHGSSLWGKGSAVHDYVTEIRIV-SPGSVNDGFAKVRVLRETTTPKEFNAA 226
Query 83 LGSEGTLGIITQVLLKVK 100
S G LG+I+QV LK++
Sbjct 227 KVSLGVLGVISQVTLKLQ 244
> At4g29740
Length=524
Score = 29.3 bits (64), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query 25 FSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPRVSSGPSLQQLMLG 84
+ +VGG ++ G + R+G V ++ V+T KG + +P+++ P L +LG
Sbjct 176 YLSVGGTLSNAGIGGQTFRHGPQISNVHELDVITGKGEM--MTCSPKLN--PELFYGVLG 231
Query 85 SEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTA 118
G GIIT+ + + P + + + F+A
Sbjct 232 GLGQFGIITRARIALDHAPTRVKWSRILYSDFSA 265
> 7301689
Length=2119
Score = 28.5 bits (62), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 32/69 (46%), Gaps = 15/69 (21%)
Query 79 QQLMLGSEGTLGIITQVLLKVKLL----PECKAYGCLAFPTFTAGVHFLRHVALDNLQPA 134
Q +M G+ GT GI+ + LL L+ PEC + F+R + L L A
Sbjct 361 QAIMKGNSGTKGILNRKLLACYLIESQNPECHSLSL-----------FMRKIVLQLLSHA 409
Query 135 SIRLMDKTQ 143
S+ D++Q
Sbjct 410 SLISRDESQ 418
> At1g23460
Length=459
Score = 28.5 bits (62), Expect = 4.5, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query 83 LGSEGTLGIITQVLLKVKLLPECKAYGCLAFPTFTAGVHFLRHVALDNLQ 132
LG + T GI+TQV+L LL E L T+ G +++ + N++
Sbjct 291 LGKDNTTGIVTQVVLDTALLRETT--NGLRIKTYQGGSGYVQGIRFTNVE 338
> YNR058w
Length=480
Score = 27.3 bits (59), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 58 TSKGSLNQQQSAPRVSSGPSLQQLMLGSEGTLGIITQVLLKVKL 101
T KG++N Q +V PSLQ L G++ + +V LK+ L
Sbjct 97 THKGAVNLVQKLLKVIDEPSLQYCFLADSGSVAV--EVALKMAL 138
> At5g56970
Length=523
Score = 27.3 bits (59), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 54/127 (42%), Gaps = 18/127 (14%)
Query 25 FSTVGGWVATRASGMKKNRYGNIEDMVVDVVVVTSKGSLNQQQSAPRVSSGPSLQQLMLG 84
+ TVGG ++ + RYG V+++ V+T KG + + L +LG
Sbjct 170 YLTVGGTLSNGGISGQTFRYGPQITNVLEMDVITGKGEI----ATCSKDMNSDLFFAVLG 225
Query 85 SEGTLGIITQVLLKVKLLPECKAYGCLAFPTF-------------TAGVHFLR-HVALDN 130
G GIIT+ +K+++ P+ + + F T GV FL + +D+
Sbjct 226 GLGQFGIITRARIKLEVAPKRAKWLRFLYIDFSEFTRDQERVISKTDGVDFLEGSIMVDH 285
Query 131 LQPASIR 137
P + R
Sbjct 286 GPPDNWR 292
Lambda K H
0.318 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40