bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1497_orf2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
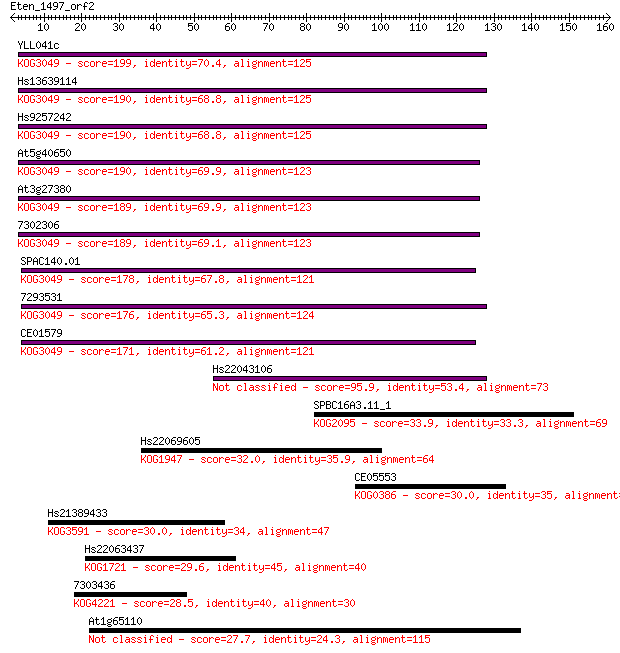
YLL041c 199 1e-51
Hs13639114 190 8e-49
Hs9257242 190 8e-49
At5g40650 190 1e-48
At3g27380 189 2e-48
7302306 189 3e-48
SPAC140.01 178 5e-45
7293531 176 1e-44
CE01579 171 6e-43
Hs22043106 95.9 3e-20
SPBC16A3.11_1 33.9 0.13
Hs22069605 32.0 0.57
CE05553 30.0 2.0
Hs21389433 30.0 2.2
Hs22063437 29.6 2.4
7303436 28.5 5.8
At1g65110 27.7 9.7
> YLL041c
Length=266
Score = 199 bits (507), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 88/125 (70%), Positives = 107/125 (85%), Gaps = 0/125 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
NFY QYKSIQP+LQR + PK+G+E+LQSIEDR KLDGLYECILCACCSTSCPSYWWN +
Sbjct 139 NFYQQYKSIQPYLQRSSFPKDGTEVLQSIEDRKKLDGLYECILCACCSTSCPSYWWNQEQ 198
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRK 122
YLGPAVLMQA+RW+ DSRD+ T+ R A +N++M LYRCH IMNC+ +CPKGL+P AI +
Sbjct 199 YLGPAVLMQAYRWLIDSRDQATKTRKAMLNNSMSLYRCHTIMNCTRTCPKGLNPGLAIAE 258
Query 123 MKKRI 127
+KK +
Sbjct 259 IKKSL 263
> Hs13639114
Length=280
Score = 190 bits (483), Expect = 8e-49, Method: Compositional matrix adjust.
Identities = 86/126 (68%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K E +EG + LQSIE+R KLDGLYECILCACCSTSCPSYWWN
Sbjct 145 NFYAQYKSIEPYLKKKDESQEGKQQYLQSIEEREKLDGLYECILCACCSTSCPSYWWNGD 204
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT ERLA++ D LYRCH IMNC+ +CPKGL+P AI
Sbjct 205 KYLGPAVLMQAYRWMIDSRDDFTEERLAKLQDPFSLYRCHTIMNCTRTCPKGLNPGKAIA 264
Query 122 KMKKRI 127
++KK +
Sbjct 265 EIKKMM 270
> Hs9257242
Length=280
Score = 190 bits (483), Expect = 8e-49, Method: Compositional matrix adjust.
Identities = 86/126 (68%), Positives = 104/126 (82%), Gaps = 1/126 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPKEGSE-ILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+P+L++K E +EG + LQSIE+R KLDGLYECILCACCSTSCPSYWWN
Sbjct 145 NFYAQYKSIEPYLKKKDESQEGKQQYLQSIEEREKLDGLYECILCACCSTSCPSYWWNGD 204
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRD+FT ERLA++ D LYRCH IMNC+ +CPKGL+P AI
Sbjct 205 KYLGPAVLMQAYRWMIDSRDDFTEERLAKLQDPFSLYRCHTIMNCTRTCPKGLNPGKAIA 264
Query 122 KMKKRI 127
++KK +
Sbjct 265 EIKKMM 270
> At5g40650
Length=280
Score = 190 bits (482), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 86/124 (69%), Positives = 101/124 (81%), Gaps = 1/124 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPK-EGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+PWL+RK G EILQS +DRAKLDG+YECILCACCSTSCPSYWWNP+
Sbjct 152 NFYNQYKSIEPWLKRKNPASVPGKEILQSKKDRAKLDGMYECILCACCSTSCPSYWWNPE 211
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
SYLGPA L+ A RWISDSRDE+T+ERL I+D KLYRCH I+NC+ +CPKGL+P I
Sbjct 212 SYLGPAALLHANRWISDSRDEYTKERLEAIDDEFKLYRCHTILNCARACPKGLNPGKQIT 271
Query 122 KMKK 125
+K+
Sbjct 272 HIKQ 275
> At3g27380
Length=279
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 86/124 (69%), Positives = 101/124 (81%), Gaps = 1/124 (0%)
Query 3 NFYTQYKSIQPWLQRKTEPK-EGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
NFY QYKSI+PWL+RKT EILQS +DRAKLDG+YECILCACCSTSCPSYWWNP+
Sbjct 153 NFYNQYKSIEPWLKRKTPASVPAKEILQSKKDRAKLDGMYECILCACCSTSCPSYWWNPE 212
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
SYLGPA L+ A RWISDSRDE+T+ERL I+D KLYRCH I+NC+ +CPKGL+P I
Sbjct 213 SYLGPAALLHANRWISDSRDEYTKERLEAIDDEFKLYRCHTILNCARACPKGLNPGKQIT 272
Query 122 KMKK 125
+K+
Sbjct 273 HIKQ 276
> 7302306
Length=297
Score = 189 bits (479), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 85/126 (67%), Positives = 102/126 (80%), Gaps = 3/126 (2%)
Query 3 NFYTQYKSIQPWLQRKTEPKEG---SEILQSIEDRAKLDGLYECILCACCSTSCPSYWWN 59
NFY QY++IQPWLQRK E E ++ LQS+EDR+KLDGLYECILCACCSTSCPSYWWN
Sbjct 152 NFYEQYRNIQPWLQRKNEAGEKKGKAQYLQSVEDRSKLDGLYECILCACCSTSCPSYWWN 211
Query 60 PQSYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGA 119
+ YLGPAVLMQA+RWI DSRDE + ERL ++ D +YRCH IMNC+ +CPKGL+P A
Sbjct 212 AEKYLGPAVLMQAYRWIIDSRDENSAERLNKLKDPFSVYRCHTIMNCTRTCPKGLNPGRA 271
Query 120 IRKMKK 125
I ++KK
Sbjct 272 IAEIKK 277
> SPAC140.01
Length=252
Score = 178 bits (451), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 82/121 (67%), Positives = 96/121 (79%), Gaps = 1/121 (0%)
Query 4 FYTQYKSIQPWLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQSY 63
FY QYKSI+PWLQ PK+ E QS DRAKLDGLYECILCACCSTSCPSYWWN + Y
Sbjct 127 FYKQYKSIEPWLQNDNIPKD-KEFYQSRADRAKLDGLYECILCACCSTSCPSYWWNSEEY 185
Query 64 LGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRKM 123
LGPAVLMQA+RWI DSRD+ T +RL + ++M +YRCH IMNC+ +CPKGL+P AI K+
Sbjct 186 LGPAVLMQAYRWIIDSRDQATAKRLDVMQNSMSVYRCHTIMNCARTCPKGLNPGLAIAKV 245
Query 124 K 124
K
Sbjct 246 K 246
> 7293531
Length=398
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 81/126 (64%), Positives = 98/126 (77%), Gaps = 2/126 (1%)
Query 4 FYTQYKSIQPWLQRKTEPKEG--SEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQ 61
FY QY+SIQPWLQRK +E ++ LQS++DR LDGLYECILCACC TSCPSYWWN
Sbjct 252 FYDQYRSIQPWLQRKDLKREAGTAQYLQSVDDRLVLDGLYECILCACCQTSCPSYWWNSN 311
Query 62 SYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIR 121
YLGPAVLMQA+RW+ DSRDE T +RL + D KLYRCH IMNC+ +CPK L+PA AI
Sbjct 312 KYLGPAVLMQAYRWVIDSRDEATEQRLDFLKDPWKLYRCHSIMNCTNTCPKHLNPARAII 371
Query 122 KMKKRI 127
++K+ +
Sbjct 372 QLKQLL 377
> CE01579
Length=298
Score = 171 bits (433), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 74/122 (60%), Positives = 96/122 (78%), Gaps = 1/122 (0%)
Query 4 FYTQYKSIQPWLQRKTEPKEGS-EILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQS 62
FY QY SIQPW+Q+KT G ++ QS+ +R +LDGLYECILCACCSTSCPSYWWN
Sbjct 160 FYAQYASIQPWIQKKTPLTLGEKQMHQSVAERDRLDGLYECILCACCSTSCPSYWWNADK 219
Query 63 YLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGLDPAGAIRK 122
YLGPAVLMQA+RW+ DSRD++ ERL +++D+ ++CH IMNC+ +CPK L+PA AI +
Sbjct 220 YLGPAVLMQAYRWVIDSRDDYATERLHRMHDSFSAFKCHTIMNCTKTCPKHLNPAKAIGE 279
Query 123 MK 124
+K
Sbjct 280 IK 281
> Hs22043106
Length=185
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 53/73 (72%), Gaps = 0/73 (0%)
Query 55 SYWWNPQSYLGPAVLMQAFRWISDSRDEFTRERLAQINDTMKLYRCHGIMNCSISCPKGL 114
SYWWN YLGPAVL+QA+ W+ DSR++FT E LA++ D + LY CH IMN + +CP GL
Sbjct 103 SYWWNRDKYLGPAVLIQAYHWMIDSREDFTEEYLAKLQDPLSLYHCHTIMNYTRTCPTGL 162
Query 115 DPAGAIRKMKKRI 127
+P A ++KK +
Sbjct 163 NPGKATAEIKKML 175
> SPBC16A3.11_1
Length=634
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 82 EFTRERLAQINDTMKLYRCHGIMNCSISCP---KGLDPAGAIRKMKKRIEEEFGTDFVKL 138
+F ++L Q N + CHGI N I+ K + A + K + EE+ ++ ++
Sbjct 310 DFLIDKLGQTNGPLVWNLCHGIDNTEITTQVQIKSMLSAKNFSQQKVKSEED-AINWFQV 368
Query 139 AAQAARSRFLEL 150
A RSRFLEL
Sbjct 369 FASDLRSRFLEL 380
> Hs22069605
Length=99
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 28/68 (41%), Gaps = 12/68 (17%)
Query 36 KLDGLYECILCACCSTSCPSYWWNPQSYLG----PAVLMQAFRWISDSRDEFTRERLAQI 91
+L GL E +L C W S LG PA+ + RWI D +D RE L
Sbjct 6 RLQGLQELVLSGCS--------WLSVSALGSAPLPALRLLDLRWIEDVKDSQLRELLLPP 57
Query 92 NDTMKLYR 99
DT R
Sbjct 58 PDTKPAER 65
> CE05553
Length=1474
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query 93 DTMKLYRCHGIM---NCSISCPKGLDPAGAIRKMKKRIEEEFG 132
D MK++ H I +IS P G+DP G +++ + I+ G
Sbjct 149 DLMKIFNLHQIRCNRPTTISVPSGIDPVGMLKQRENMIQNRIG 191
> Hs21389433
Length=160
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 1/48 (2%)
Query 11 IQP-WLQRKTEPKEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYW 57
+QP WL+R + P G + D+ +GL E L A C T+ Y+
Sbjct 7 VQPSWLRRASAPLPGLSAPGRLFDQRFGEGLLEAELAALCPTTLAPYY 54
> Hs22063437
Length=229
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 6/44 (13%)
Query 21 PKEGSE--ILQSIEDRAKLDGLYECILCACCS--TSCPSYWWNP 60
P EG + +LQ D L GL C+L ACC CP+ W P
Sbjct 10 PSEGLQWHLLQLSADAVSLLGLGHCVLSACCPGLAQCPA--WRP 51
> 7303436
Length=1526
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 18 KTEPKEGSEILQSIEDRAKLDGLYECILCA 47
+T+ K GS + S+E+ L G Y+C+L A
Sbjct 96 RTQLKNGSLYISSVEENRGLTGAYQCLLTA 125
> At1g65110
Length=1094
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 54/116 (46%), Gaps = 20/116 (17%)
Query 22 KEGSEILQSIEDRAKLDGLYECILCACCSTSCPSYWWNPQSYLGPAVLMQAFRWISDSRD 81
+EG + L+ + D A+++ ++ +C CS Q++ P + D++
Sbjct 182 REGLDALEQVLDSARINRKWKFWMCRTCS----------QTFFYPKKFKNHLEQVHDAKY 231
Query 82 EFTRERLAQ-INDTMKLYRCHGIMNCSISCPKGLDPAGAIRKMKKRIEEEFGTDFV 136
+ RE LAQ I+D G++ S++ + +D A +K R+ EF +FV
Sbjct 232 KPVREDLAQSIDDVWA-----GMI--SVADWEPVDALAAAEMIKNRL--EFVKEFV 278
Lambda K H
0.321 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40