bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1495_orf2
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
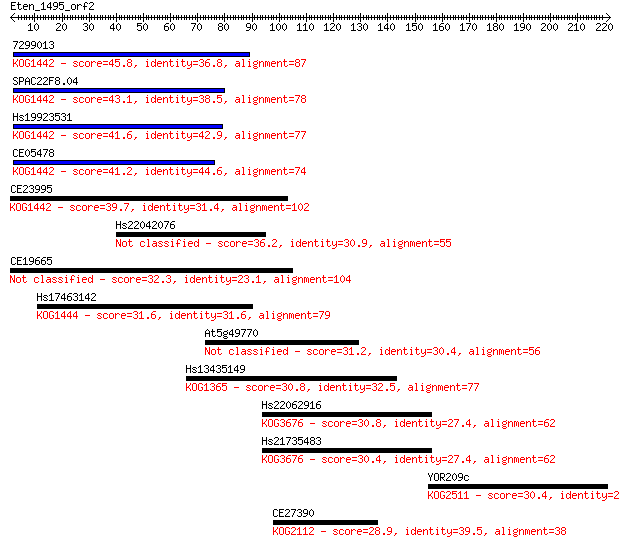
7299013 45.8 6e-05
SPAC22F8.04 43.1 4e-04
Hs19923531 41.6 0.001
CE05478 41.2 0.002
CE23995 39.7 0.004
Hs22042076 36.2 0.047
CE19665 32.3 0.86
Hs17463142 31.6 1.4
At5g49770 31.2 1.7
Hs13435149 30.8 2.3
Hs22062916 30.8 2.5
Hs21735483 30.4 2.6
YOR209c 30.4 3.1
CE27390 28.9 7.5
> 7299013
Length=337
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 51/93 (54%), Gaps = 10/93 (10%)
Query 2 VQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSDTASVT-- 59
V V+ Y + RSLT VFS++L+ L+Q+ + C + +GF L G D S+T
Sbjct 116 VTVAFYYIGRSLTTVFSVVLTYVILRQRTSFKCLLCCGAIVVGFWL----GVDQESLTEV 171
Query 60 ----GYIMGALASLFQATYTVQMKATLVFIEKE 88
G I G L+SL A +++Q K +L ++ +E
Sbjct 172 FSWRGTIFGVLSSLALAMFSIQTKKSLGYVNQE 204
> SPAC22F8.04
Length=383
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 40/78 (51%), Gaps = 1/78 (1%)
Query 2 VQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSDTASVTGY 61
V VS YQ++R L L F++LLS LKQK CL V +GF S A + G
Sbjct 175 VPVSFYQISRGLLLPFTILLSFVLLKQKTRLFPFGGCLLVMLGFGFGVRFESHVAPI-GI 233
Query 62 IMGALASLFQATYTVQMK 79
I+G +S A +V +K
Sbjct 234 ILGVWSSFTTAIESVAVK 251
> Hs19923531
Length=351
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 42/86 (48%), Gaps = 13/86 (15%)
Query 2 VQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSD------T 55
V V+ Y V RSLT VF++LLS LKQ + +C + GF L G D T
Sbjct 125 VGVAFYNVGRSLTTVFNVLLSYLLLKQTTSFYALLTCGIIIGGFWL----GVDQEGAEGT 180
Query 56 ASVTGYIMGALASL---FQATYTVQM 78
S G + G LASL A YT ++
Sbjct 181 LSWLGTVFGVLASLCVSLNAIYTTKV 206
> CE05478
Length=416
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 39/83 (46%), Gaps = 13/83 (15%)
Query 2 VQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSD------T 55
V VS Y V RSLT VF+++ + L QK + C + GF L G D T
Sbjct 177 VGVSFYYVGRSLTTVFNVVCTYLILGQKTSGQAIGCCALIIFGFLL----GVDQEGVTGT 232
Query 56 ASVTGYIMGALASL---FQATYT 75
S TG I G LASL A YT
Sbjct 233 LSYTGVIFGVLASLSVALNAIYT 255
> CE23995
Length=332
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 52/108 (48%), Gaps = 10/108 (9%)
Query 1 RVQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSDTAS--- 57
V V+ Y V RS+T VF+++L+ + + SCL + +GF +GSD S
Sbjct 123 NVGVAFYYVGRSITTVFTVILTYVFFGDNSTKGVNVSCLVILIGF----GIGSDQESQDP 178
Query 58 --VTGYIMGALASLFQATYTVQMKA-TLVFIEKESDTLLPAAKPRSAY 102
+G + G ASL A + K+ T+V ++K + L KP +
Sbjct 179 LTTSGVLYGMFASLAVALNALYTKSNTMVLVQKFNKQLSQTIKPNKYF 226
> Hs22042076
Length=393
Score = 36.2 bits (82), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 40 FVAMGFALMTAVGSDTASVTGYIMGALASLFQATYTVQMKATLVFIEKESDTLLP 94
F+ +AL+T V ++ +A LFQA Q+ L KE D++LP
Sbjct 301 FLCRAYALLTVVSKCGINIYSMPFAQMALLFQAIQAFQLSCDLAVTPKEGDSILP 355
> CE19665
Length=647
Score = 32.3 bits (72), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 42/104 (40%), Gaps = 6/104 (5%)
Query 1 RVQVSVYQVARSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSDTASVTG 60
R + + + +V L LS F L Q + +C+ M + M + S+ +
Sbjct 504 RFFIMAFSMGNFAAIVMYLTLSAFHLNQIATYCFIINCIMYLMYYGCMKVLHSERITSKA 563
Query 61 YIMGALASLFQATYTVQMKATLVFIEKESDTLLPAAKPRSAYRP 104
+ GAL+ L A A F + ++D AA R+ +P
Sbjct 564 KLCGALSLLAWAV------AGFFFFQDDTDWTRSAAASRALNKP 601
> Hs17463142
Length=416
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 34/79 (43%), Gaps = 8/79 (10%)
Query 11 RSLTLVFSLLLSVFWLKQKVVRAEVFSCLFVAMGFALMTAVGSDTASVTGYIMGALASLF 70
R L LV ++L+ V LK V + + + A + G T GY+ G LA L
Sbjct 109 RCLPLV-TMLIGVLVLKNGAPSPGVLAAVLITTCGAALAGAGDLTGDPIGYVTGVLAVLV 167
Query 71 QATYTVQMKATLVFIEKES 89
A Y LV I+K S
Sbjct 168 HAAY-------LVLIQKAS 179
> At5g49770
Length=946
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 24/56 (42%), Gaps = 0/56 (0%)
Query 73 TYTVQMKATLVFIEKESDTLLPAAKPRSAYRPITNENAATAQAYSDVTGLDPNPDS 128
T +Q L EK D L +P RP +E ++ + GL+PN DS
Sbjct 864 TTIIQNSGNLKGFEKYVDVALQCVEPEGVNRPTMSEVVQELESILRLVGLNPNADS 919
> Hs13435149
Length=727
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 34/82 (41%), Gaps = 5/82 (6%)
Query 66 LASLFQATYTV-QMKATLVFIEK----ESDTLLPAAKPRSAYRPITNENAATAQAYSDVT 120
L L TYT Q TL+ E S LLPAA+ +A P+ Q Y + T
Sbjct 579 LPCLSPPTYTTFQATPTLIPTETAALYPSSALLPAARVPAAPTPVAYYPGPATQLYLNYT 638
Query 121 GLDPNPDSQDSDGGIFSKPRSA 142
P+P + G + P +A
Sbjct 639 AYYPSPPVSPTTVGYLTTPTAA 660
> Hs22062916
Length=735
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 26/62 (41%), Gaps = 1/62 (1%)
Query 94 PAAKPRSAYRPITNENAATAQAYSDVTGLDPNPDSQDSDGGIFSKPRSATIDMKLPAELR 153
PA P IT +A + ++ G +PNP + +FSKP + I +
Sbjct 23 PAILPEKRPAEIT-PTKKSAHFFLEIEGFEPNPTVAKTSPPVFSKPMDSNIRQCISGNCD 81
Query 154 DM 155
DM
Sbjct 82 DM 83
> Hs21735483
Length=790
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 26/62 (41%), Gaps = 1/62 (1%)
Query 94 PAAKPRSAYRPITNENAATAQAYSDVTGLDPNPDSQDSDGGIFSKPRSATIDMKLPAELR 153
PA P IT +A + ++ G +PNP + +FSKP + I +
Sbjct 23 PAILPEKRPAEIT-PTKKSAHFFLEIEGFEPNPTVAKTSPPVFSKPMDSNIRQCISGNCD 81
Query 154 DM 155
DM
Sbjct 82 DM 83
> YOR209c
Length=429
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/66 (22%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 155 MAETNWDYEAALQEGQEEGQQPKDDDLELFQEGQQRHSRPMRLQNLAEREGLPTINGGAA 214
+ +WDYE L++ +++ + D+ + F E R R ++ Q+L + + +NG
Sbjct 146 FVDIDWDYENQLEQAEKKAETLFDNGIR-FSEFGTRRRRSLKAQDLIMQGIMKAVNGNPD 204
Query 215 KSSAGL 220
++ + L
Sbjct 205 RNKSLL 210
> CE27390
Length=223
Score = 28.9 bits (63), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query 98 PRSAYRPITNENAATAQAYSDVTGLDPNPDSQDSDGGI 135
P S+ RP+T A+ D+ GLDPN +Q+ + GI
Sbjct 53 PHSSERPVTLNMGMRMPAWFDLFGLDPN--AQEDEQGI 88
Lambda K H
0.315 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4183546302
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40