bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
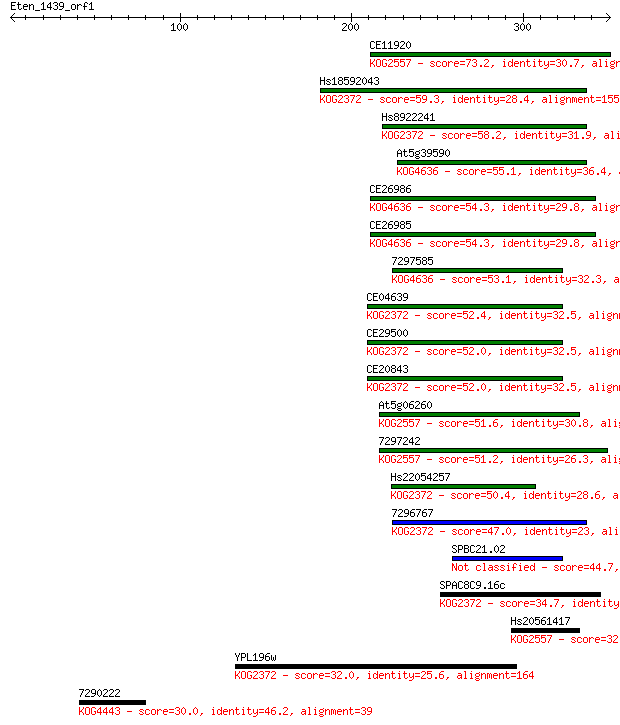
Query= Eten_1439_orf1
Length=350
Score E
Sequences producing significant alignments: (Bits) Value
CE11920 73.2 7e-13
Hs18592043 59.3 1e-08
Hs8922241 58.2 3e-08
At5g39590 55.1 2e-07
CE26986 54.3 4e-07
CE26985 54.3 4e-07
7297585 53.1 9e-07
CE04639 52.4 2e-06
CE29500 52.0 2e-06
CE20843 52.0 2e-06
At5g06260 51.6 2e-06
7297242 51.2 3e-06
Hs22054257 50.4 6e-06
7296767 47.0 5e-05
SPBC21.02 44.7 3e-04
SPAC8C9.16c 34.7 0.35
Hs20561417 32.7 1.2
YPL196w 32.0 2.1
7290222 30.0 6.9
> CE11920
Length=399
Score = 73.2 bits (178), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 71/140 (50%), Gaps = 2/140 (1%)
Query 211 MTSTMFPFPPKAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSE 270
+ ++ PF + WT LY++ KHG SFS+L C P + V+++ +G+ G +S+
Sbjct 187 VLNSHLPFDRRKNWTLLYSNMKHGQSFSQL-VKCINGEGPCMIVIRSMKGRRFG-FFASQ 244
Query 271 LKEGGHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRF 330
G + G A LF L P++ +G N+VYLN + + HP GLG GG +
Sbjct 245 GFLAGPQYRGTAECFLFQLAPKIATFDATGRTENYVYLNYQQQQHPNGLGIGGTESVWPL 304
Query 331 WIGDEMKDCYITKSDCTYSP 350
+I +E K+ ++ P
Sbjct 305 FIHEEFGGGTCQKNSSSFEP 324
> Hs18592043
Length=197
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 74/158 (46%), Gaps = 15/158 (9%)
Query 182 SEETKGWHGQGTLDARSRCFTDEHAFVLRMTSTMFPFPPKA---PWTCLYASWKHGTSFS 238
+E++ HG+ + D R R + +L F FPP+ PW+ ++ + + G S
Sbjct 19 AEDSLTAHGRLSRDRRMRGLRWRYTRLLS-----FHFPPRVTGHPWSLVFCTSRDGFSLQ 73
Query 239 RLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEGGHNFFGDANTCLFSLEPQLNVLRT 298
L S P++ V++ ++GQ+ GA SS ++ F+G T LFS PQL V +
Sbjct 74 SLYRRMEGCSGPVLLVLRDQDGQIFGAFSSSAIRL-SKGFYGTGETFLFSFSPQLKVFKW 132
Query 299 SGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRFWIGDEM 336
+G FV K + +G G G F W+ ++
Sbjct 133 TGSNSFFV----KGDLDSLMMGSGS--GRFGLWLDGDL 164
> Hs8922241
Length=269
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 58/122 (47%), Gaps = 10/122 (8%)
Query 218 FPPKA---PWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEG 274
PP+ PWT +Y + KHGTS L P++ V+K +GQV GA+ S LK
Sbjct 122 LPPRTIGYPWTLVYGTGKHGTSLKTLYRTMTGLDTPVLMVIKDSDGQVFGALASEPLK-V 180
Query 275 GHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRFWIGD 334
F+G T +F+ P+ V + +G F+ + + L FGG G F W+
Sbjct 181 SDGFYGTGETFVFTFCPEFEVFKWTGDDMFFIKGDMDS------LAFGGGGGEFALWLDG 234
Query 335 EM 336
++
Sbjct 235 DL 236
> At5g39590
Length=542
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 65/131 (49%), Gaps = 26/131 (19%)
Query 227 LYASWKHGTSFSRLCANCFFYSAPMVAVV-----------KTEEGQVLGAIISSELKEGG 275
LY S+ HG +RL +N Y AP++ ++ +E V+GAI L++G
Sbjct 320 LYRSYYHGKGMNRLWSNVEGYHAPILVIISASCKVEHEATSSERKWVIGAI----LQQGF 375
Query 276 HN---FFGDANTCLFSLEPQLNVLRTSGLGRNFVY--LNTKNKFH-----PIGLGFGGQV 325
N F+G + LFS+ P + +SG +NF Y L+ + P+G+GFGG +
Sbjct 376 ENRDAFYGSSGN-LFSISPVFHAFSSSGKEKNFAYSHLHPAGGVYDAHPKPVGIGFGGTL 434
Query 326 GAFRFWIGDEM 336
G R +I ++
Sbjct 435 GNERIFIDEDF 445
> CE26986
Length=411
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 62/136 (45%), Gaps = 12/136 (8%)
Query 211 MTSTMFPFPP-----KAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGA 265
+ + FP P ++ WT LY S +HG S +R F Y P V + + ++G+V+
Sbjct 200 LPAVYFPAKPTETSGESHWTPLYTSLQHGISTNRFETLVFDYRGPTVTIFRMKDGRVVVI 259
Query 266 IISSELKEGGHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQV 325
E + G N FG T F + P N+ R G N +Y N K + GL F ++
Sbjct 260 AADQEWRHSG-NRFGGTFTSFFEIVP--NIRRIDGA--NSIYCNLKLRSSAYGLSFKNEL 314
Query 326 GAFRFWIGDEMKDCYI 341
+ + DE+ D +
Sbjct 315 KIEKDF--DEILDIEV 328
> CE26985
Length=415
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 62/136 (45%), Gaps = 12/136 (8%)
Query 211 MTSTMFPFPP-----KAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGA 265
+ + FP P ++ WT LY S +HG S +R F Y P V + + ++G+V+
Sbjct 204 LPAVYFPAKPTETSGESHWTPLYTSLQHGISTNRFETLVFDYRGPTVTIFRMKDGRVVVI 263
Query 266 IISSELKEGGHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQV 325
E + G N FG T F + P N+ R G N +Y N K + GL F ++
Sbjct 264 AADQEWRHSG-NRFGGTFTSFFEIVP--NIRRIDGA--NSIYCNLKLRSSAYGLSFKNEL 318
Query 326 GAFRFWIGDEMKDCYI 341
+ + DE+ D +
Sbjct 319 KIEKDF--DEILDIEV 332
> 7297585
Length=526
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Query 224 WTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEGGHNFFGDAN 283
WT LY S +HG +R + Y P + ++ T++ Q SE KE H F G
Sbjct 346 WTLLYNSNEHGVGANRFLHHVLGYRGPTLVLLHTKDEQTYCVASPSEWKE-THLFVGGEG 404
Query 284 TCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFG 322
+C+ L P+ +L N +YLNT + +P GL G
Sbjct 405 SCVIQLLPKFVILEKKP---NILYLNTSIRGYPKGLRAG 440
> CE04639
Length=536
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 59/125 (47%), Gaps = 12/125 (9%)
Query 209 LRMTSTMFPFPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQV 262
L + M PP+A PW +Y S KHG S + + + +P++ +++ + V
Sbjct 368 LMIRQVMDILPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHV 427
Query 263 LGAIISSELKEGGHNFFGDANTCL---FSLE-PQLNVLRTSG-LGRNFVYLNTKNKFHPI 317
GA++SS ++ H FFG ++CL F+ E P LR G N ++N I
Sbjct 428 FGAVVSSAIRPNDH-FFGTGDSCLLWRFTGEVPHTRELRQYNWTGDNQYFVNAAKDSLSI 486
Query 318 GLGFG 322
G G G
Sbjct 487 GAGSG 491
> CE29500
Length=817
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 59/125 (47%), Gaps = 12/125 (9%)
Query 209 LRMTSTMFPFPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQV 262
L + M PP+A PW +Y S KHG S + + + +P++ +++ + V
Sbjct 649 LMIRQVMDILPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHV 708
Query 263 LGAIISSELKEGGHNFFGDANTCL---FSLE-PQLNVLRTSG-LGRNFVYLNTKNKFHPI 317
GA++SS ++ H FFG ++CL F+ E P LR G N ++N I
Sbjct 709 FGAVVSSAIRPNDH-FFGTGDSCLLWRFTGEVPHTRELRQYNWTGDNQYFVNAAKDSLSI 767
Query 318 GLGFG 322
G G G
Sbjct 768 GAGSG 772
> CE20843
Length=838
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 59/125 (47%), Gaps = 12/125 (9%)
Query 209 LRMTSTMFPFPPKA---PWTCLYASWKHGTSFSRLCANCFFYS---APMVAVVKTEEGQV 262
L + M PP+A PW +Y S KHG S + + + +P++ +++ + V
Sbjct 670 LMIRQVMDILPPRAEGYPWVNIYNSEKHGFSLATMYRKMAEFDEDLSPVLLIIRDTKEHV 729
Query 263 LGAIISSELKEGGHNFFGDANTCL---FSLE-PQLNVLRTSG-LGRNFVYLNTKNKFHPI 317
GA++SS ++ H FFG ++CL F+ E P LR G N ++N I
Sbjct 730 FGAVVSSAIRPNDH-FFGTGDSCLLWRFTGEVPHTRELRQYNWTGDNQYFVNAAKDSLSI 788
Query 318 GLGFG 322
G G G
Sbjct 789 GAGSG 793
> At5g06260
Length=424
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 56/120 (46%), Gaps = 4/120 (3%)
Query 216 FPFPPKAPWTCLYASWKHGTSFSRLCANCFFYS-APMVAVVKTEEGQVLGAIISSELKEG 274
P W LY S HG SF+ + + V ++K EG V G +S+ E
Sbjct 230 LPHHELVEWKLLYHSSVHGQSFNTFLGHTSNTGMSASVLIIKDTEGYVYGGY-ASQPWER 288
Query 275 GHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVY--LNTKNKFHPIGLGFGGQVGAFRFWI 332
+F+GD + LF L P+ + R +G N + N ++ P G+GFGG++ F +I
Sbjct 289 YSDFYGDMKSFLFQLNPKAAIYRPTGANTNIQWCATNFTSENIPNGIGFGGKINHFGLFI 348
> 7297242
Length=448
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 57/133 (42%), Gaps = 2/133 (1%)
Query 216 FPFPPKAPWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEGG 275
P + W L++S +G SFS + P + ++ E+ + G +SE
Sbjct 247 LPREHRHKWRFLFSSKINGESFSTMLGKVL-DKGPTLFFIEDEDQYIFGGY-ASETWSVK 304
Query 276 HNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRFWIGDE 335
F GD ++ L++L P + + ++ YLN + P GLG GGQ + WI
Sbjct 305 PQFGGDDSSLLYTLSPAMRCFSATTYNNHYQYLNLNQQTMPNGLGMGGQFDFWGLWIDCS 364
Query 336 MKDCYITKSDCTY 348
D +S TY
Sbjct 365 FGDGQSVESCTTY 377
> Hs22054257
Length=942
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Query 223 PWTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEGGHNFFGDA 282
PW Y++ +HGTS L +P++ V+K + Q+ GA + K H ++G
Sbjct 803 PWRLAYSTLEHGTSLKTLYRKSASLDSPVLLVIKDMDNQIFGAYATHPFKFSDH-YYGTG 861
Query 283 NTCLFSLEPQLNVLRTSGLGRNFV 306
T L++ P V + SG F+
Sbjct 862 ETFLYTFSPHFKVFKWSGENSYFI 885
> 7296767
Length=1325
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/113 (23%), Positives = 50/113 (44%), Gaps = 7/113 (6%)
Query 224 WTCLYASWKHGTSFSRLCANCFFYSAPMVAVVKTEEGQVLGAIISSELKEGGHNFFGDAN 283
W+ ++++ +HG + + L +P++ V++ E V GA+ S L H F+G
Sbjct 1187 WSLIFSTSQHGFALNSLYRKMARLESPVLIVIEDTEHNVFGALTSCSLHVSDH-FYGTGE 1245
Query 284 TCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRFWIGDEM 336
+ L+ P V +G F+ N ++ L G G F W+ ++
Sbjct 1246 SLLYKFNPSFKVFHWTGENMYFIKGNMES------LSIGAGDGRFGLWLDGDL 1292
> SPBC21.02
Length=511
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 36/64 (56%), Gaps = 5/64 (7%)
Query 259 EGQVLGAIISSELKEGGHNFFGDANTCLFSLEPQLNVLRTSGLGRNFVYLNTKNKFHPIG 318
E +LGA IS+ ++ FFGD +T LF L+P V S L +N+ + KN +G
Sbjct 377 ENILLGAYISTRWRQSHMGFFGDHSTLLFQLQPIHQVYYASNLDKNYCMFD-KN----VG 431
Query 319 LGFG 322
LGFG
Sbjct 432 LGFG 435
> SPAC8C9.16c
Length=188
Score = 34.7 bits (78), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 40/94 (42%), Gaps = 7/94 (7%)
Query 252 VAVVKTEEGQVLGAIISSELKEGGHNFFGDANTCLFSLEPQLNVLRTSGLGR-NFVYLNT 310
+ V+ +G V G I L H +FG T L+ P + +G NFV T
Sbjct 72 ILAVRDTDGDVFGVFIPDYLIPAPH-YFGSEETFLWKYFPPKKYVHYPFVGNSNFVAYCT 130
Query 311 KNKFHPIGLGFGGQVGAFRFWIGDEMKDCYITKS 344
K+ L FGG G + W+ ++ Y +++
Sbjct 131 KS-----FLAFGGGNGRYSLWLDGSLEYAYSSRT 159
> Hs20561417
Length=124
Score = 32.7 bits (73), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 293 LNVLRTSGLGRNFVYLNTKNKFHPIGLGFGGQVGAFRFWI 332
+ V +G +++YLN + P GLG GGQ F W+
Sbjct 1 MAVYTHTGYNDHYMYLNHGQQTIPNGLGMGGQHNYFGLWV 40
> YPL196w
Length=273
Score = 32.0 bits (71), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 42/177 (23%), Positives = 70/177 (39%), Gaps = 16/177 (9%)
Query 132 VQYTVYAARRRHEGHSSIRESTRGTADSSSFGRGVSKSPAWKRSTSAEAISEE--TKGWH 189
V+ ++ +R G S + TA SS S +P ++TS I EE
Sbjct 4 VKDAIFKIKRSIAGTDSSDSTAYTTASESSPQLKDSHNPFRNKTTSERTIVEEGSLPPVR 63
Query 190 GQGTLDA-RSRCFTDEHAFVLRMTSTMFP--FPPKAPWTCLYASWKHGTSFSRLCANCF- 245
G L + +++ T E +R T+ P W LY+ +HG+S L +N
Sbjct 64 LNGYLPSTKNKLLTPEMCDEIR---TLMPTRIQLYTEWNLLYSLEQHGSSLHSLYSNVAP 120
Query 246 ----FYSAPMVAVVKTEEGQVLGAIISSELKEGGH-NFFGDANTCLFSLE--PQLNV 295
F V V+K + + GA + H + G+ L+ L+ P +N+
Sbjct 121 DSKEFRRVGYVLVIKDRKNGIFGAYSNEAFHPNEHRQYTGNGECFLWKLDKVPDVNI 177
> 7290222
Length=2304
Score = 30.0 bits (66), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 3/39 (7%)
Query 41 SIGNGTGSGTRRGTSSTELAAQASGAILHASSGTIGSGG 79
S+GN TG+GT GT + A S IL+A++GT G G
Sbjct 162 SVGNSTGTGTPAGTP---IGATTSTIILNANNGTAGVSG 197
Lambda K H
0.319 0.132 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8481321646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40