bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1438_orf2
Length=174
Score E
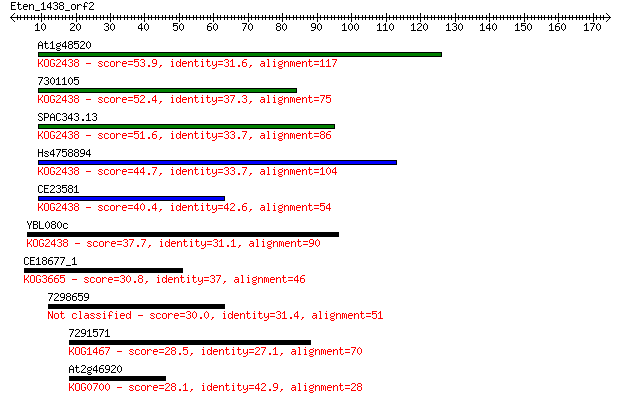
Sequences producing significant alignments: (Bits) Value
At1g48520 53.9 2e-07
7301105 52.4 5e-07
SPAC343.13 51.6 7e-07
Hs4758894 44.7 1e-04
CE23581 40.4 0.002
YBL080c 37.7 0.011
CE18677_1 30.8 1.6
7298659 30.0 2.2
7291571 28.5 7.6
At2g46920 28.1 9.6
> At1g48520
Length=550
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 37/120 (30%), Positives = 62/120 (51%), Gaps = 5/120 (4%)
Query 9 RVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQ--EIQPETRHWDDALKRSVPLRSKVN-S 65
+VEIKN+ + AI RA+ E RQ + +G+ +I ETR W++ +++V +R K +
Sbjct 281 KVEIKNLNAFSAISRAIDFEISRQALLYNQGKADQIVTETRLWEEGAQKTVTMRKKEGLA 340
Query 66 SWVFFPFEETQIPPVSLDKETKARIKRGLVLSAASRRSLYRQWGLELSLRTAIARDIKAA 125
+ +FP E +P V L +E I+ L ++R Y GL + +A D+ A
Sbjct 341 DYRYFP--EPDLPEVILTQEYVDSIRASLPELPEAKRRRYEAMGLGMQDVLFLANDVSVA 398
> 7301105
Length=467
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 45/76 (59%), Gaps = 3/76 (3%)
Query 9 RVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSK-VNSSW 67
R E+KN+ S+R+I +A+ E RQ + + G I ETR+WD +R+V +R K V +
Sbjct 234 RTEVKNIGSVRSISQAITYEINRQLETVANGGVITNETRNWDAENRRTVAMRDKEVLQDY 293
Query 68 VFFPFEETQIPPVSLD 83
F P E +PP+ ++
Sbjct 294 RFMP--EPNLPPLHVN 307
> SPAC343.13
Length=526
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 47/87 (54%), Gaps = 3/87 (3%)
Query 9 RVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSKVNS-SW 67
RVE+KN+ ++R + ++ E RQ + G ETR +DD ++ PLR+K S +
Sbjct 243 RVELKNLSNVRNVVNSIRHEVDRQVSLANMGVSWPSETRGFDDITGKTFPLRNKTTSDDY 302
Query 68 VFFPFEETQIPPVSLDKETKARIKRGL 94
+F P ET IPP+ L + + + L
Sbjct 303 LFLP--ETDIPPIILSRSYVNSVLKSL 327
> Hs4758894
Length=557
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 58/119 (48%), Gaps = 17/119 (14%)
Query 9 RVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSKVN-SSW 67
R E+KN+ S+R + +A+ E +RQ + +E G EI ETR + L ++ +R K +
Sbjct 276 RTEVKNLNSIRFLAKAIDYEIQRQINELENGGEILNETRSFHHKLGCTMSMRDKEGKQDY 335
Query 68 VFFPFEETQIPPVSLDKETKA-------------RIKRGL-VLSAASRRSLYRQWGLEL 112
F P E +PP+ L T +I+ L L + +R L +Q+G+ L
Sbjct 336 RFMP--EPNLPPLVLYDATSLPAGADPQQVINIDQIRETLPELPSVTREKLVQQYGMLL 392
> CE23581
Length=350
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 33/54 (61%), Gaps = 1/54 (1%)
Query 9 RVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSK 62
R EIKN+ S+R I A++ E RQ +I++ G +Q ETR D R+V +R K
Sbjct 79 RTEIKNMNSIRTIHTAINYEIARQFEILKNGGAVQNETRAADSE-GRTVSMRDK 131
> YBL080c
Length=541
Score = 37.7 bits (86), Expect = 0.011, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 54/94 (57%), Gaps = 9/94 (9%)
Query 6 DFPRVEIKNVRSLRAIRRALHAEDKRQKDIIEKGQE---IQPETRHWDDALKRSVPLRSK 62
++ RVE+KN+ + +I A+ E +RQ ++I G ++PETR W + +V LRSK
Sbjct 235 EYARVELKNLPNTSSIINAIKYEYQRQVELISVGDTSSLMEPETRGWTGS--STVKLRSK 292
Query 63 VNS-SWVFFPFEETQIPPVSLDKETKARIKRGLV 95
+ + + P + ++P ++L + + + RGL+
Sbjct 293 ETTIDYRYMP--DPELPYINLAPDVISGV-RGLM 323
> CE18677_1
Length=631
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 9/55 (16%)
Query 5 RDFPRVEIKNVR-SLRAIRRALHAEDKRQ--------KDIIEKGQEIQPETRHWD 50
RD + I N + +LR + R + E+ +Q +DI + E++PE R WD
Sbjct 431 RDMCMINILNCKETLRILHRGMREEEVKQTMSDVDGTRDIHRRLTELRPEGRKWD 485
> 7298659
Length=472
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 12 IKNVRSLRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSK 62
+K+V +L + R LH D R++D+ E G Q D R +R+K
Sbjct 41 VKSVTNLVPVARYLHKFDVREQDVSEMGASFQTLVDRLDHINSREHCIRTK 91
> 7291571
Length=626
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 18 LRAIRRALHAEDKRQKDIIEKGQEIQPETRHWDDALKRSVPLRSKVNSSWVFFPFEETQI 77
L A+R+ +H + K E G+ + +H D L + PL V++++ F + TQ+
Sbjct 334 LHALRQVVHDFETPAKK--EFGRSLDAAVKHHVDHLHKCRPLAVSVSNAYKQFKNQLTQL 391
Query 78 PPVSLDKETK 87
P + E+K
Sbjct 392 PADVPETESK 401
> At2g46920
Length=814
Score = 28.1 bits (61), Expect = 9.6, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 18 LRAIRRALHAEDKRQKDIIEKGQEIQPE 45
LRA+ RAL + ++ D++EK +I PE
Sbjct 541 LRAMARALESTEEAYMDMVEKSLDINPE 568
Lambda K H
0.319 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2634976318
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40