bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1428_orf1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
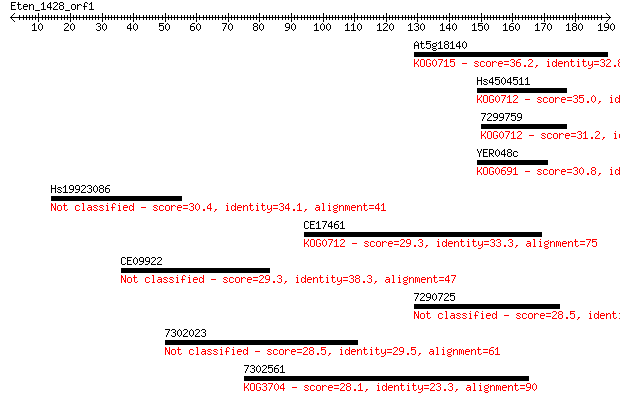
At5g18140 36.2 0.043
Hs4504511 35.0 0.10
7299759 31.2 1.3
YER048c 30.8 1.7
Hs19923086 30.4 2.1
CE17461 29.3 4.9
CE09922 29.3 5.4
7290725 28.5 7.7
7302023 28.5 7.8
7302561 28.1 9.8
> At5g18140
Length=333
Score = 36.2 bits (82), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 37/61 (60%), Gaps = 4/61 (6%)
Query 129 RAYSSSSSSSATATTATGGGSHYETLGVKPNATQAEIRQARRDTKALKAERHEFDAGTDR 188
R+ ++ +S++ ++++ TGG +HY LG+ NATQ +I++A R L A + D D
Sbjct 57 RSKTTITSAAFSSSSNTGGQNHYAVLGIARNATQGDIKRAYR----LLARKFHPDVNKDS 112
Query 189 R 189
+
Sbjct 113 K 113
> Hs4504511
Length=397
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 1/28 (3%)
Query 149 SHYETLGVKPNATQAEIRQARRDTKALK 176
++Y+ LGVKPNATQ E+++A R ALK
Sbjct 6 TYYDVLGVKPNATQEELKKAYRKL-ALK 32
> 7299759
Length=403
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 20/27 (74%), Gaps = 1/27 (3%)
Query 150 HYETLGVKPNATQAEIRQARRDTKALK 176
+Y+ LGVKPNAT E+++A R ALK
Sbjct 7 YYDILGVKPNATPDELKKAYRKL-ALK 32
> YER048c
Length=391
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 149 SHYETLGVKPNATQAEIRQARR 170
+Y+ LG+KP AT EI++A R
Sbjct 6 EYYDILGIKPEATPTEIKKAYR 27
> Hs19923086
Length=212
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Query 14 GGIQDLGSDFHRHVRCYCSSGCDRNIHSRIGRRGFDKGYNW 54
GG QD+ S R +CS C +N+H + G+++G +W
Sbjct 129 GGSQDMSSAVKR----FCSVSCLQNLHVLESKDGWERGRSW 165
> CE17461
Length=374
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 11/81 (13%)
Query 94 DLTMSSSLCRREGVWW-FSACSARRLFVVGASC-----SVFRAYSSSSSSSATATTATGG 147
DLT S+ R + FS + R LF + C V R SS T
Sbjct 41 DLTRVCSISRFSAFFIQFSLINVRCLFTKFSFCFHLLVGVVRCNMFGGGSSGPVDTTL-- 98
Query 148 GSHYETLGVKPNATQAEIRQA 168
Y TL V+P+A+QA+I+++
Sbjct 99 ---YTTLNVRPDASQADIKKS 116
> CE09922
Length=739
Score = 29.3 bits (64), Expect = 5.4, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 36 DRNIHSRIGRRGFDKGYNWSLSSSNYSTGIGSHHALSYRRSSNSNVI 82
D + S G DK YN S Y +GIG + L Y S S VI
Sbjct 413 DVQVGSVDGLTTLDKKYNKDTKSDAYLSGIGRYLNLKYTGSKTSEVI 459
> 7290725
Length=2951
Score = 28.5 bits (62), Expect = 7.7, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 129 RAYSSSSSSSATATTATGGGSHYET--LGVKPNAT-QAEIRQARRDTKA 174
RA+++S SS+ A + T GSH++T V N T + E+R+ D++A
Sbjct 713 RAFAASLRSSSPADSTTSVGSHHQTPRSSVSSNRTFRREMREGSHDSQA 761
> 7302023
Length=901
Score = 28.5 bits (62), Expect = 7.8, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Query 50 KGYNWSLSSSNYSTGIGSH-HALSYRRSSNSNVIGGGGSEGGSTSDLTMSSSLCRREGVW 108
+G+N +S + G SH HA+ + +N I EGG+ L ++ S+ E V
Sbjct 568 QGHNVRISGEDVGRGTFSHRHAMLVDQQTNEMFIPLNSMEGGNGGKLELAHSILSEEAVL 627
Query 109 WF 110
F
Sbjct 628 GF 629
> 7302561
Length=298
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 38/90 (42%), Gaps = 14/90 (15%)
Query 75 RSSNSNVIGGGGSEGGSTSDLTMSSSLCRREGVWWFSACSARRLFVVGASCSVFRAYSSS 134
R ++ ++GG G+ T T+SSSL RR+ + + + ++
Sbjct 100 RGYDNALLGGSGAGAKETKGKTVSSSLVRRQALGSGGGVAGAAM--------------TT 145
Query 135 SSSSATATTATGGGSHYETLGVKPNATQAE 164
++ S A+ A +E L + PN T E
Sbjct 146 TTMSPMASAAQMAAKSFEELAIFPNGTVNE 175
Lambda K H
0.316 0.128 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3166472848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40