bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1426_orf1
Length=157
Score E
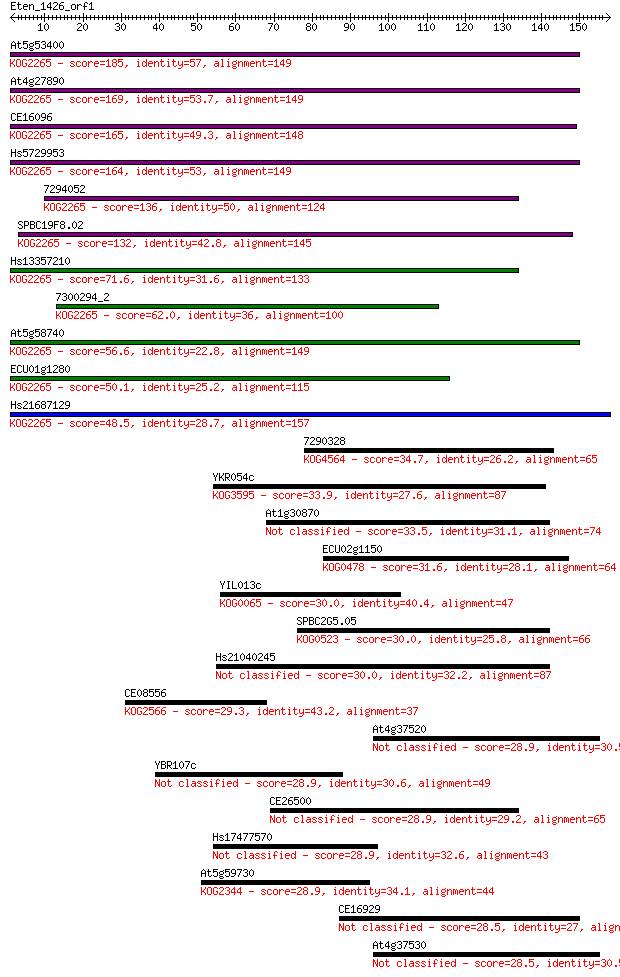
Sequences producing significant alignments: (Bits) Value
At5g53400 185 3e-47
At4g27890 169 2e-42
CE16096 165 3e-41
Hs5729953 164 7e-41
7294052 136 2e-32
SPBC19F8.02 132 2e-31
Hs13357210 71.6 7e-13
7300294_2 62.0 4e-10
At5g58740 56.6 2e-08
ECU01g1280 50.1 2e-06
Hs21687129 48.5 5e-06
7290328 34.7 0.078
YKR054c 33.9 0.13
At1g30870 33.5 0.20
ECU02g1150 31.6 0.71
YIL013c 30.0 2.0
SPBC2G5.05 30.0 2.0
Hs21040245 30.0 2.0
CE08556 29.3 3.4
At4g37520 28.9 3.9
YBR107c 28.9 4.3
CE26500 28.9 4.5
Hs17477570 28.9 4.6
At5g59730 28.9 4.7
CE16929 28.5 5.5
At4g37530 28.5 6.3
> At5g53400
Length=304
Score = 185 bits (470), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 85/149 (57%), Positives = 109/149 (73%), Gaps = 2/149 (1%)
Query 1 VDVYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVH 60
V++ VP GT R C +K +RL VGLKGQ PI+DGE + V DDC W +EDQ +
Sbjct 158 VNIPVPTGTKARTVVCEIK--KNRLKVGLKGQDPIVDGELYRSVKPDDCYWNIEDQKVIS 215
Query 61 LSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLP 120
+ L K D+M WW C +KG+PEIDT+ + PE SKL DLDPETR+TVEKMM+DQ+QKQ GLP
Sbjct 216 ILLTKSDQMEWWKCCVKGEPEIDTQKVEPETSKLGDLDPETRSTVEKMMFDQRQKQMGLP 275
Query 121 TSDQQRQAEMLEKFKQSHPELDFSNAKIN 149
TS++ ++ E+L+KF HPE+DFSNAK N
Sbjct 276 TSEELQKQEILKKFMSEHPEMDFSNAKFN 304
> At4g27890
Length=293
Score = 169 bits (428), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 80/149 (53%), Positives = 105/149 (70%), Gaps = 2/149 (1%)
Query 1 VDVYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVH 60
+++ +P GT R+ C +K +RL VGLKGQ I+DGEF V DDC W +EDQ +
Sbjct 147 INIPMPEGTKSRSVTCEIK--KNRLKVGLKGQDLIVDGEFFNSVKPDDCFWNIEDQKMIS 204
Query 61 LSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLP 120
+ L K D+M WW +KG+PEIDT+ + PE SKL DLDPETRA+VEKMM+DQ+QKQ GLP
Sbjct 205 VLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLDPETRASVEKMMFDQRQKQMGLP 264
Query 121 TSDQQRQAEMLEKFKQSHPELDFSNAKIN 149
SD+ + +ML+KF +P +DFSNAK N
Sbjct 265 RSDEIEKKDMLKKFMAQNPGMDFSNAKFN 293
> CE16096
Length=320
Score = 165 bits (418), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 109/150 (72%), Gaps = 2/150 (1%)
Query 1 VDVYVPVGTG--VRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNT 58
++V +P+ G +++ VK+ + ++VGLK Q PI+DG+ + ++C W +E+
Sbjct 170 LEVKIPIAAGFAIKSRDVVVKIEKTSVSVGLKNQAPIVDGKLPHAIKVENCNWVIENGKA 229
Query 59 VHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRG 118
+ L+LEK++ M WW+ L DP I+TK + PENSKLSDLD ETRA VEKMMYDQ+QK+ G
Sbjct 230 IVLTLEKINDMEWWNRFLDSDPPINTKEVKPENSKLSDLDGETRAMVEKMMYDQRQKEMG 289
Query 119 LPTSDQQRQAEMLEKFKQSHPELDFSNAKI 148
LPTSD++++ +ML++F + HPE+DFSNAKI
Sbjct 290 LPTSDEKKKHDMLQQFMKQHPEMDFSNAKI 319
> Hs5729953
Length=331
Score = 164 bits (415), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 79/151 (52%), Positives = 104/151 (68%), Gaps = 2/151 (1%)
Query 1 VDVYVP--VGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNT 58
+D+ VP V ++ V + L VGLKGQP I+DGE +V ++ W +ED
Sbjct 181 LDLAVPFCVNFRLKGKDMVVDIQRRHLRVGLKGQPAIIDGELYNEVKVEESSWLIEDGKV 240
Query 59 VHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRG 118
V + LEK+++M WWS ++ DPEI+TK I PENSKLSDLD ETR+ VEKMMYDQ+QK G
Sbjct 241 VTVHLEKINKMEWWSRLVSSDPEINTKKINPENSKLSDLDSETRSMVEKMMYDQRQKSMG 300
Query 119 LPTSDQQRQAEMLEKFKQSHPELDFSNAKIN 149
LPTSD+Q++ E+L+KF HPE+DFS AK N
Sbjct 301 LPTSDEQKKQEILKKFMDQHPEMDFSKAKFN 331
> 7294052
Length=332
Score = 136 bits (342), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 62/124 (50%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 10 GVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVHLSLEKVDRM 69
G+RA + + L VG+KGQ PI+DGE +V ++ +W L+D TV ++L+K+++M
Sbjct 209 GLRARDLVISIGKKSLKVGIKGQTPIIDGELCGEVKTEESVWVLQDSKTVMITLDKINKM 268
Query 70 RWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAE 129
WWS ++ DPEI T+ I PE+SKLSDLD ETR+ VEKMMYDQ+QK+ GLPTS+ +++ +
Sbjct 269 NWWSRLVTTDPEISTRKINPESSKLSDLDGETRSMVEKMMYDQRQKELGLPTSEDRKKQD 328
Query 130 MLEK 133
+LEK
Sbjct 329 ILEK 332
> SPBC19F8.02
Length=166
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/145 (42%), Positives = 97/145 (66%), Gaps = 5/145 (3%)
Query 3 VYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVHLS 62
++VP GT ++ Q + ++ + + + +L G ++++ D+ WT+E+Q + +
Sbjct 24 IHVPKGTRAKSLQVDMSNHDLKIQINVPERKVLLSGPLEKQINLDESTWTVEEQERLVIH 83
Query 63 LEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTS 122
LEK ++M WWSCV+KG P ID +I PENSKLSDLD ETRATVEKMM +Q QK+ +
Sbjct 84 LEKSNKMEWWSCVIKGHPSIDIGSIEPENSKLSDLDEETRATVEKMMLEQSQKR-----T 138
Query 123 DQQRQAEMLEKFKQSHPELDFSNAK 147
D+Q++ ++L+ F + HPELDFSN +
Sbjct 139 DEQKRKDVLQNFMKQHPELDFSNVR 163
> Hs13357210
Length=220
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 79/137 (57%), Gaps = 6/137 (4%)
Query 1 VDVYVPVGTGV-RAAQCSVKVTGSRLTVGL---KGQPPILDGEFSQKVHADDCMWTLEDQ 56
++V VPV V + Q SV ++ S + V + G+ +++G+ + K++ + +W+LE
Sbjct 58 LEVRVPVPKHVVKGKQVSVALSSSSIRVAMLEENGERVLMEGKLTHKINTESSLWSLEPG 117
Query 57 NTVHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQ 116
V ++L KV WW+ +L+G+ ID I E S ++ +D E +A ++++ +D QK
Sbjct 118 KCVLVNLSKVGEY-WWNAILEGEEPIDIDKINKERS-MATVDEEEQAVLDRLTFDYHQKL 175
Query 117 RGLPTSDQQRQAEMLEK 133
+G P S + + EML+K
Sbjct 176 QGKPQSHELKVHEMLKK 192
> 7300294_2
Length=188
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 57/102 (55%), Gaps = 5/102 (4%)
Query 13 AAQCSVKVTGSRLTVGLKGQPP--ILDGEFSQKVHADDCMWTLEDQNTVHLSLEKVDRMR 70
A + + + + V K P IL+G SQ++ + +WT+ DQN + +S +K +
Sbjct 42 AKKLHISIQAQHIKVSSKHSPETIILEGNLSQRIKHKEAVWTI-DQNRLIISYDKAKEL- 99
Query 71 WWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQ 112
WW + +GDPEID+K I E + DL ET+AT+EK+ Q
Sbjct 100 WWDRLFEGDPEIDSKKIECERY-IDDLPEETQATIEKLRVQQ 140
> At5g58740
Length=158
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/149 (22%), Positives = 66/149 (44%), Gaps = 36/149 (24%)
Query 1 VDVYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVH 60
V++Y+ + V K+ + VG+KG PP L+ + S V D WTLED + +H
Sbjct 30 VNMYITLPPNVHPKSFHCKIQSKHIEVGIKGNPPYLNHDLSAPVKTDCSFWTLED-DIMH 88
Query 61 LSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLP 120
++L+K ++ + W+ + G ++D
Sbjct 89 ITLQKREKGQTWASPILGQGQLDPY----------------------------------- 113
Query 121 TSDQQRQAEMLEKFKQSHPELDFSNAKIN 149
+D +++ ML++F++ +P DFS A+ +
Sbjct 114 ATDLEQKRLMLQRFQEENPGFDFSQAQFS 142
> ECU01g1280
Length=131
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 60/116 (51%), Gaps = 4/116 (3%)
Query 1 VDVYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVH 60
+++ PV ++ +++ G ++ V +G+ ++DGE +V W + + + V
Sbjct 16 INIQFPVTGDADSSAIKIRIVGKKICVKNQGEI-VIDGELLHEVDVSSLWWVI-NGDVVD 73
Query 61 LSLEKVDRMRWWSCVLKGDPEIDTKTIVP-ENSKLSDLDPETRATVEKMMYDQQQK 115
+++ K R WW +L G +D + + +++ +S LD E R VEKMM++ K
Sbjct 74 VNVTK-KRNEWWDSLLVGSESVDVQKLAENKHADMSMLDAEAREVVEKMMHNTSGK 128
> Hs21687129
Length=157
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 45/159 (28%), Positives = 67/159 (42%), Gaps = 41/159 (25%)
Query 1 VDVYVPVGTGVRAAQCSVKVTGSRLTVGLKGQPPILDGEFSQKVHADDCMWTLEDQNTVH 60
++V VP GT + QC ++ L+VG + IL G+ AD+ WTLED+ V
Sbjct 30 IEVQVPPGTRAQDIQCGLQSRHVALSVGGR---EILKGKLFDSTIADEGTWTLEDRKMVR 86
Query 61 LSLEKVDR--MRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRG 118
+ L K R W+ +L+ + D + Q Q QR
Sbjct 87 IVLTKTKRDAANCWTSLLESEYAADP-------------------------WVQDQMQRK 121
Query 119 LPTSDQQRQAEMLEKFKQSHPELDFSNAKINWGNSGWGG 157
L LE+F++ +P DFS A+I+ GN GG
Sbjct 122 L----------TLERFQKENPGFDFSGAEIS-GNYTKGG 149
> 7290328
Length=633
Score = 34.7 bits (78), Expect = 0.078, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query 78 GDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEMLEKFKQS 137
GDP K I P N +L+ P + + V M+++ QQ+QR +P + + + ++ +++
Sbjct 514 GDPSATGKRISPPNKRLNGRKPSSASIV--MIHEPQQRQRLMPRLQNKAREKGKDRVEKT 571
Query 138 HPELD 142
E +
Sbjct 572 DAEAE 576
> YKR054c
Length=4092
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 51/87 (58%), Gaps = 13/87 (14%)
Query 54 EDQNTVHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQ 113
E+Q V++ LEK++ VLK + E++ KT+ ++++L++ + E R+T++KM+ +Q
Sbjct 3012 ENQRFVNVGLEKLNE-----SVLKVN-ELN-KTLSKKSTELTEKEKEARSTLDKMLMEQN 3064
Query 114 QKQRGLPTSDQQRQAEMLEKFKQSHPE 140
+ +R +Q E ++K + E
Sbjct 3065 ESER------KQEATEEIKKILKVQEE 3085
> At1g30870
Length=349
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 11/85 (12%)
Query 68 RMRWWSCVLKGDPEIDTKTI------VPENSKLSDLDPETRATVEKMMYDQQQKQRGLPT 121
R+ ++ DP ID K S+ DLDP T A + Y QK G+ +
Sbjct 226 RLYNYNATSGSDPSIDAKYADYLQRRCRWASETVDLDPVTPAVFDNQYYINLQKHMGVLS 285
Query 122 SDQQ-----RQAEMLEKFKQSHPEL 141
+DQ+ R A +++ F + P++
Sbjct 286 TDQELVKDPRTAPLVKTFAEQSPQI 310
> ECU02g1150
Length=708
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 6/64 (9%)
Query 83 DTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEMLEKFKQSHPELD 142
+ + IVP L PE+ + + D +Q G + RQ E L + ++H +
Sbjct 547 EARRIVPR------LTPESMKMLTQSYVDLRQMDNGKTITATTRQLESLIRLSEAHARMR 600
Query 143 FSNA 146
FSNA
Sbjct 601 FSNA 604
> YIL013c
Length=1411
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 24/53 (45%), Gaps = 6/53 (11%)
Query 56 QNTVHLSLEKVDRMRWWSCVLKGDPEIDTKTIV------PENSKLSDLDPETR 102
Q VHL L V SCVL+GD + D +V P ++DL P R
Sbjct 830 QQDVHLELLTVRESLEISCVLRGDGDRDYLGVVSNLLRLPSEKLVADLSPTQR 882
> SPBC2G5.05
Length=685
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 33/70 (47%), Gaps = 4/70 (5%)
Query 76 LKGDPEIDTKTIVPEN----SKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEML 131
L+G + + PE+ KL DP V +Y +++ + +S ++ +M
Sbjct 261 LQGTHSVHGSPLKPEDCVHVKKLFGFDPTKTFQVPPEVYAYYKERVAIASSAEEEYKKMY 320
Query 132 EKFKQSHPEL 141
+KQS+P+L
Sbjct 321 ASYKQSYPDL 330
> Hs21040245
Length=445
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 42/90 (46%), Gaps = 9/90 (10%)
Query 55 DQNTVHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKLSDLD---PETRATVEKMMYD 111
+Q +V L E DR + W + EI K + +SDL+ E A +E+M D
Sbjct 94 EQISVTLGDEMFDRKKRW------ESEIPDKGRFSRTNIISDLEEQISELTAIIEQMNRD 147
Query 112 QQQKQRGLPTSDQQRQAEMLEKFKQSHPEL 141
Q Q+ L + R AEM + F+ + EL
Sbjct 148 HQSAQKLLSSEMDLRCAEMKQNFENKNREL 177
> CE08556
Length=561
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 4/38 (10%)
Query 31 GQPPILDGEFSQKVHA-DDCMWTLEDQNTVHLSLEKVD 67
G+ PI +FS +V+ DD LED N H SL++ D
Sbjct 184 GRVPIASNDFSSRVYTYDD---NLEDYNMAHFSLQRED 218
> At4g37520
Length=329
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 11/59 (18%)
Query 96 DLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEMLEKFKQSHPELDFSNAKINWGNSG 154
++DP T + + Y Q+ +GL TSDQ +L ++S P +D W N+G
Sbjct 247 NMDPTTPRQFDNVYYKNLQQGKGLFTSDQ-----VLFTDRRSKPTVDL------WANNG 294
> YBR107c
Length=245
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 26/49 (53%), Gaps = 5/49 (10%)
Query 39 EFSQKVHADDCMWTLEDQNTVHLSLEKVDRMRWWSCVLKGDPEIDTKTI 87
+F QK M L D + VH++ E ++++ W+S L +PE K +
Sbjct 115 KFEQK-----TMIKLADASIVHVTKENIEQITWFSSKLYFEPETQDKNL 158
> CE26500
Length=668
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 6/65 (9%)
Query 69 MRWWSCVLKGDPEIDTKTIVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQA 128
+ +W LK D +D+K ++ DL E R + K++ +Q T+D R+A
Sbjct 177 LEFW---LKTDQNLDSKA---KSLIFEDLSLENRPKIGKLIALEQALTEAQDTNDMYREA 230
Query 129 EMLEK 133
+LEK
Sbjct 231 GILEK 235
> Hs17477570
Length=1361
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 54 EDQNTVHLSLE-KVDRMRWWSCVLKGDPEIDTKTIVPENSKLSD 96
ED + L L+ + +R ++ C +KG I K ++P+N+ +SD
Sbjct 1133 EDSGLLFLKLKAQNEREKFAFCSMKGCERIKIKALIPKNAGVSD 1176
> At5g59730
Length=634
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 22/44 (50%), Gaps = 1/44 (2%)
Query 51 WTLEDQNTVHLSLEKVDRMRWWSCVLKGDPEIDTKTIVPENSKL 94
W + V L +K +++ W VL PEI T I PE +K+
Sbjct 476 WVFRHEEKVKLYADKFEKLAWGK-VLDLLPEIPTDEISPEEAKV 518
> CE16929
Length=340
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 31/63 (49%), Gaps = 4/63 (6%)
Query 87 IVPENSKLSDLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEMLEKFKQSHPELDFSNA 146
+V E +SD+ P R +E+M+ + R P D+ + EK +++H E D +
Sbjct 68 LVREKMIISDIAPNDRRKIERML----KNLRPPPVFDEFLTEDETEKVQKAHSERDVDSV 123
Query 147 KIN 149
+N
Sbjct 124 LMN 126
> At4g37530
Length=329
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 11/59 (18%)
Query 96 DLDPETRATVEKMMYDQQQKQRGLPTSDQQRQAEMLEKFKQSHPELDFSNAKINWGNSG 154
++DP T + + Y Q+ +GL TSDQ +L +S P +D W N+G
Sbjct 247 NMDPNTPRQFDNVYYKNLQQGKGLFTSDQ-----VLFTDSRSKPTVDL------WANNG 294
Lambda K H
0.314 0.131 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40