bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1395_orf1
Length=107
Score E
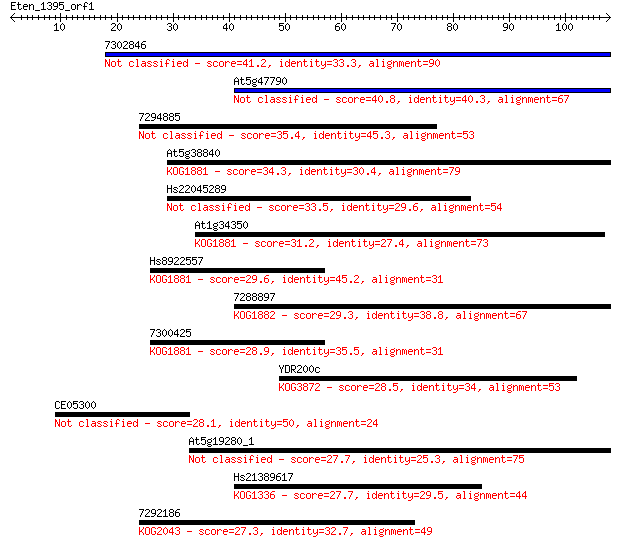
Sequences producing significant alignments: (Bits) Value
7302846 41.2 5e-04
At5g47790 40.8 5e-04
7294885 35.4 0.024
At5g38840 34.3 0.055
Hs22045289 33.5 0.11
At1g34350 31.2 0.45
Hs8922557 29.6 1.3
7288897 29.3 1.9
7300425 28.9 2.1
YDR200c 28.5 3.1
CE05300 28.1 3.6
At5g19280_1 27.7 4.8
Hs21389617 27.7 5.5
7292186 27.3 6.9
> 7302846
Length=383
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 48/92 (52%), Gaps = 3/92 (3%)
Query 18 DATTVEILLRGKSFYRIGKLAD-NDLVALNPSVSRRHAALVVLKGGHV-LLIDLKSKART 75
D ++++ K Y G+ + ND + S SR H+A V K ++ L+DL S T
Sbjct 26 DKLVQKLMVDEKRCYLFGRNSQMNDFCIDHASCSRVHSAFVYHKHLNIAYLVDLGSTHGT 85
Query 76 FKNGMPLDHDHVGVQMQTNDSFSLGASSRHYL 107
F + L+ H Q+Q N +F GAS+R+Y+
Sbjct 86 FIGTLRLE-AHKPTQLQINSTFHFGASTRNYI 116
> At5g47790
Length=369
Score = 40.8 bits (94), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 41 DLVALNPSVSRRHAALVVLKGGHVLLIDLKSKARTFKNGMPLDHDHVGVQMQTNDSFSLG 100
D V + SVSR+HAA+V K G + +IDL S TF L D V+++ S
Sbjct 97 DFVLDHQSVSRQHAAVVPHKNGSIFVIDLGSAHGTFVANERLTKD-TPVELEVGQSLRFA 155
Query 101 ASSRHYL 107
AS+R YL
Sbjct 156 ASTRIYL 162
> 7294885
Length=811
Score = 35.4 bits (80), Expect = 0.024, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Query 24 ILLRGKSFYRIGKLADNDLVALNPSVSRRHAALVV---LKGGHVLLI-DLKSKARTF 76
+L GK Y IG+LA + +VA + S+SR HA L++ G L I DL S+ TF
Sbjct 26 VLFPGKKVYTIGRLATDLIVAQDLSISRNHAQLLIQTEADGDDTLHIEDLGSRYGTF 82
> At5g38840
Length=730
Score = 34.3 bits (77), Expect = 0.055, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 34/79 (43%), Gaps = 1/79 (1%)
Query 29 KSFYRIGKLADNDLVALNPSVSRRHAALVVLKGGHVLLIDLKSKARTFKNGMPLDHDHVG 88
K Y G+ D +PS+SR HA + + G + DL S T N +D V
Sbjct 117 KGAYLFGRDGICDFALEHPSISRFHAVIQYKRSGAAYIFDLGSTHGTTVNKNKVD-KKVF 175
Query 89 VQMQTNDSFSLGASSRHYL 107
V + D G S+R Y+
Sbjct 176 VDLNVGDVIRFGGSTRLYI 194
> Hs22045289
Length=123
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 26/54 (48%), Gaps = 6/54 (11%)
Query 29 KSFYRIGKLADNDLVALNPSVSRRHAALVVLKGGHVLLIDLKSKARTFKNGMPL 82
+SF+R G+ A P+ +RRH V+ GGH + + + RT +P
Sbjct 17 RSFWRFGRSAKR------PAGTRRHTYTVIPTGGHTYMFAVYTVTRTTPTQIPC 64
> At1g34350
Length=1587
Score = 31.2 bits (69), Expect = 0.45, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 39/74 (52%), Gaps = 2/74 (2%)
Query 34 IGKLADNDLVALNPSVSRRHAALVVLKGGHVLLI-DLKSKARTFKNGMPLDHDHVGVQMQ 92
+G+ D D++ +PS+SR H + + L + DL S T+ + ++ H V+++
Sbjct 176 VGRHPDCDILLTHPSISRFHLEIRSISSRQKLFVTDLSSVHGTWVRDLRIE-PHGCVEVE 234
Query 93 TNDSFSLGASSRHY 106
D+ +G S+R Y
Sbjct 235 EGDTIRIGGSTRIY 248
> Hs8922557
Length=796
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 26 LRGKSFYRIGKLADNDLVALNPSVSRRHAAL 56
L+G S+ G+L+ D+ +PSVSR HA L
Sbjct 183 LKGTSYCLFGRLSGCDVCLEHPSVSRYHAVL 213
> 7288897
Length=421
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 34/79 (43%), Gaps = 13/79 (16%)
Query 41 DLVALNPSVSRRHAALVVL-------KGGH-----VLLIDLKSKARTFKNGMPLDHDHVG 88
DL +PS S++HAAL G H + LIDL S TF N +D
Sbjct 313 DLAVDHPSCSKQHAALQYRLVPFEREDGSHGKRVRLYLIDLDSANGTFLNNKKIDARKY- 371
Query 89 VQMQTNDSFSLGASSRHYL 107
++ D G SSR Y+
Sbjct 372 YELIEKDVIKFGFSSREYV 390
> 7300425
Length=726
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 26 LRGKSFYRIGKLADNDLVALNPSVSRRHAAL 56
L+ ++ + G+L +ND+ A +P++SR H L
Sbjct 82 LQQQAVWTFGRLPENDVPAAHPTISRFHVVL 112
> YDR200c
Length=604
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 3/54 (5%)
Query 49 VSRRHAALVV-LKGGHVLLIDLKSKARTFKNGMPLDHDHVGVQMQTNDSFSLGA 101
+SR HA L G + + DLKS TF NG+ + + V+++ D+ LG
Sbjct 223 LSRNHACLSCDPTSGKIYIRDLKSSNGTFVNGVKIRQN--DVELKVGDTVDLGT 274
> CE05300
Length=586
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 9 LRLLERREGDATTVEILLRGKSFY 32
L+ +E EGD T IL RG FY
Sbjct 348 LKFIELEEGDTTKSAILFRGTIFY 371
> At5g19280_1
Length=295
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 41/83 (49%), Gaps = 8/83 (9%)
Query 33 RIGKLADNDLVALNPSVSRRHAALVVLKGGHVL-LIDLKSKARTFKNGMPLDHDHVG--- 88
++G+++ +DL + VS +HA + L+D+ S T N + H +G
Sbjct 209 KLGRVSPSDLALKDSEVSGKHAQITWNSTKFKWELVDMGSLNGTLVNSHSISHPDLGSRK 268
Query 89 ----VQMQTNDSFSLGASSRHYL 107
V++ ++D +LG +++ Y+
Sbjct 269 WGNPVELASDDIITLGTTTKVYV 291
> Hs21389617
Length=605
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 41 DLVALNPSVSRRHAALVVLKGGHVLLIDLKSKARTFKNGMPLDH 84
+ +AL P R + VL V+ +D+++K FK+G L++
Sbjct 247 EQLALRPKEFFRAYGIEVLTEAQVVTVDVRTKKVVFKDGFKLEY 290
> 7292186
Length=1259
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 24 ILLRGKSFYRIGKLADNDLVALNPSVSRRHAALVVLKGGHVLLIDLKSK 72
ILL+ + YRIG+ ++ + S+ HA +L+ G V L L K
Sbjct 14 ILLKADTIYRIGRQKGLEISIADESMELAHATACILRRGVVRLAALVGK 62
Lambda K H
0.320 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40