bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1337_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
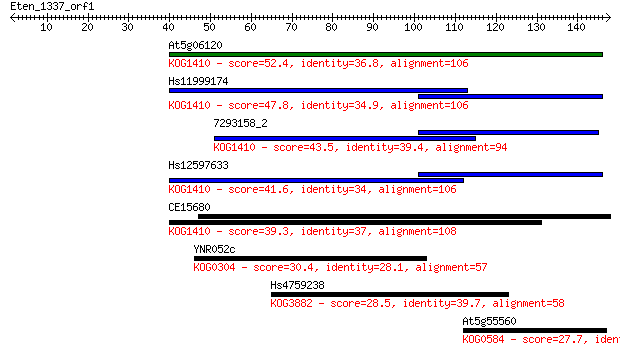
At5g06120 52.4 3e-07
Hs11999174 47.8 7e-06
7293158_2 43.5 2e-04
Hs12597633 41.6 5e-04
CE15680 39.3 0.003
YNR052c 30.4 1.3
Hs4759238 28.5 5.1
At5g55560 27.7 8.3
> At5g06120
Length=1059
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 57/109 (52%), Gaps = 12/109 (11%)
Query 40 ESPLNNEVLRQEQLEVLSQLARCRYTKTA---RMVHDLFEETKAKAQSGEMNRKVFEEKI 96
ESPL+ E L E L L+ + R +T A + L TK Q+G K +
Sbjct 258 ESPLSKEAL--ECLVRLASVRRSLFTNDATRSNFLAHLMTGTKEILQTG----KGLADHD 311
Query 97 TWLVYHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQ 145
YH FCRL+G+ ++ LSEL E + E+ Q + +FT++SL+SWQ
Sbjct 312 N---YHVFCRLLGRFRLNYQLSELVKMEGYGEWIQLVAEFTLKSLQSWQ 357
> Hs11999174
Length=1087
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 7/80 (8%)
Query 40 ESPLNNEVLRQEQLEVLSQLARCRYTKTARMVHDLFEETK-------AKAQSGEMNRKVF 92
E PL + L Q+QL+ LS + RC Y KT ++ LF+++ A + M+ V
Sbjct 430 EDPLEDTGLVQQQLDQLSTIGRCEYEKTCALLVQLFDQSAQSYQELLQSASASPMDIAVQ 489
Query 93 EEKITWLVYHEFCRLIGKVN 112
E ++TWLVY + G+V+
Sbjct 490 EGRLTWLVYIIGAVIGGRVS 509
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 101 YHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQ 145
YHEFCRL+ ++ +++ L EL E + E + + +FT+ SL+ W+
Sbjct 327 YHEFCRLLARLKSNYQLGELVKVENYPEVIRLIANFTVTSLQHWE 371
> 7293158_2
Length=500
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 101 YHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSW 144
YHEFCRL+ ++ +++ L EL A + E Q + FT+ESL W
Sbjct 250 YHEFCRLLARLKSNYQLGELIAVPCYPEAIQLIAKFTVESLHLW 293
Score = 34.3 bits (77), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query 51 EQLEVLSQLARCRYTKTARMVHDLFEETKAK--------AQSGEMNRKVFEEKITWLVYH 102
+QLE LS + RC Y KT ++ F++ KA+ + ++ + E ++TWLVY
Sbjct 346 QQLEQLSVIERCEYNKTCNLLVQHFDQ-KAREYENLLQTPNANSIDITIHELQLTWLVYI 404
Query 103 EFCRLIGKVNTS 114
++G++ +
Sbjct 405 IGSAIVGRLTVA 416
> Hs12597633
Length=1088
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 101 YHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQ 145
YHEFCR + ++ T++ L EL + + E + + +FT+ SL+ W+
Sbjct 326 YHEFCRFLARLKTNYQLGELVMVKEYPEVIRLIANFTITSLQHWE 370
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 12/79 (15%)
Query 40 ESPLNNEVLRQEQLEVLSQLARCRYTKTARMVHDLFEETKAKAQ------SG-EMNRKVF 92
+ PL++ +QLE L ++RC Y KT ++ LF++ Q SG ++ +
Sbjct 429 DDPLDDTATVFQQLEQLCTVSRCEYEKTCALLVQLFDQNAQNYQKLLHPYSGVTVDITIQ 488
Query 93 EEKITWLVYHEFCRLIGKV 111
E ++ WLVY L+G V
Sbjct 489 EGRLAWLVY-----LVGTV 502
> CE15680
Length=1078
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 59/129 (45%), Gaps = 40/129 (31%)
Query 47 VLRQEQLE----VLSQLARCRYTKTARMVHDLFEETKAKAQSGEMNRKVFEEKITWLV-- 100
VL QE E +++QLA R T LF T+ +A +K+ E ++ ++
Sbjct 274 VLPQESSEKVMTIIAQLASIRRT--------LFNGTERQAYV----QKLVEGVVSVIMNP 321
Query 101 --------YHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKS--------- 143
+HEFCRLI ++ T++ L EL A + + L +FT++SL+
Sbjct 322 GKLSDQAAFHEFCRLIARLKTNYQLCELIAVPCYSHMLRLLAEFTVQSLRMMEFSANSTY 381
Query 144 -----WQRM 147
WQRM
Sbjct 382 FLMTFWQRM 390
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 4/95 (4%)
Query 40 ESPLNNEVLRQEQLEVLSQLARCRYTKTARMVHDLFEETKAKAQSG---EMNRKVFEEKI 96
E+PL+++ + +E L+ + RC Y T +++ F++ +G + N + E ++
Sbjct 433 ENPLDDQGATLQVMEHLAIICRCEYENTCKLLTQHFDQNANIWMNGSENDANTAIAEGRL 492
Query 97 TWLVYHEFCRLIGKVN-TSHHLSELCASEPFREFP 130
WL+ + GK TS + + E FP
Sbjct 493 VWLITLIGTAVFGKTTATSSDVHDKMDGELIARFP 527
> YNR052c
Length=433
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 46 EVLRQEQLEVLSQLARCRYTKTARMVHDLFEETKAKAQSGEMNRKVFEEKITWLVYH 102
E++ E LE+L + + + K + D+FE ++ SG M ++ +TW+ YH
Sbjct 255 EIMSTESLELLRK-SGINFEKHENLGIDVFEFSQLLMDSGLM----MDDSVTWITYH 306
> Hs4759238
Length=237
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 23/66 (34%), Positives = 37/66 (56%), Gaps = 11/66 (16%)
Query 65 TKTARMVHD-LFEETKAKAQSGEMNRKVFEEKITWLVYHEFCRLIGKVNTS-------HH 116
+K+ R+V++ L+E TK + +GE + K F+E I +V+ E + G VN + H
Sbjct 111 SKSDRIVNETLYENTKLLSATGE-SEKQFQEAI--IVFQEEFKCCGLVNGAADWGNNFQH 167
Query 117 LSELCA 122
ELCA
Sbjct 168 YPELCA 173
> At5g55560
Length=313
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 112 NTSHHLSELCASEPFREFPQHLFDFTMESLKSWQR 146
NT + ++E+C S RE+ + +M +LK W +
Sbjct 104 NTLNFITEICTSGNLREYRKKHRHVSMRALKKWSK 138
Lambda K H
0.322 0.131 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40