bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1312_orf1
Length=183
Score E
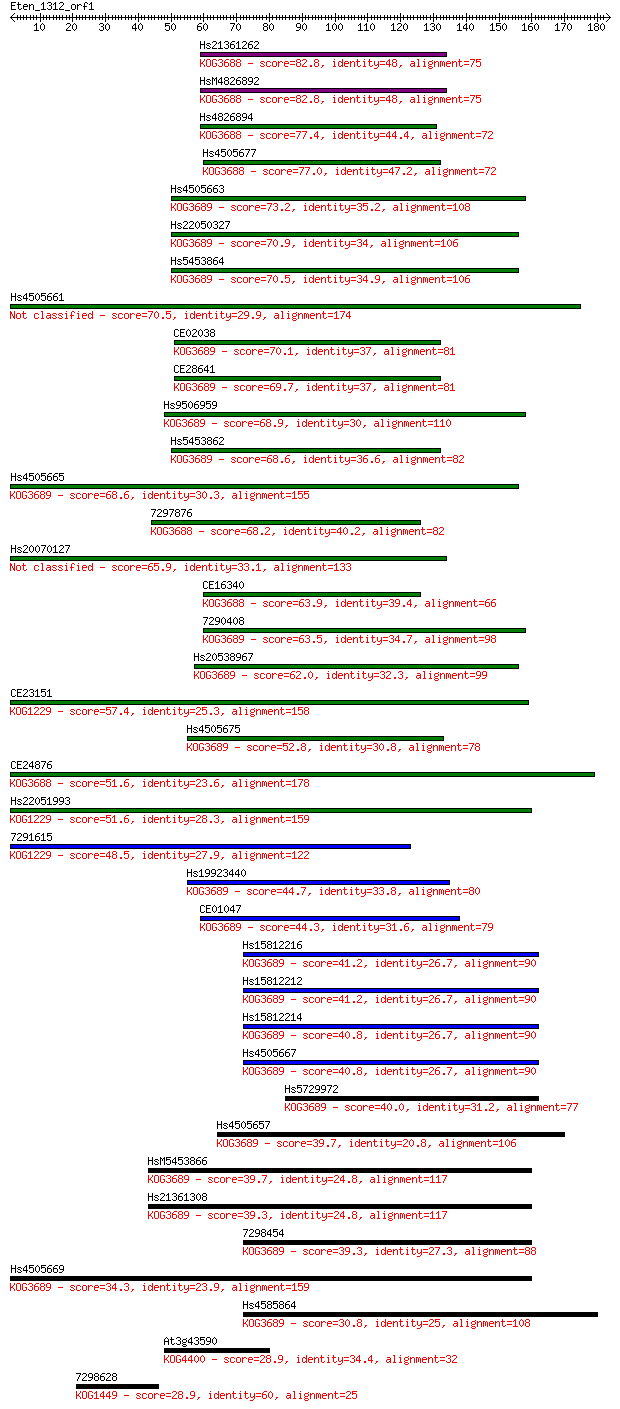
Sequences producing significant alignments: (Bits) Value
Hs21361262 82.8 4e-16
HsM4826892 82.8 4e-16
Hs4826894 77.4 2e-14
Hs4505677 77.0 2e-14
Hs4505663 73.2 3e-13
Hs22050327 70.9 1e-12
Hs5453864 70.5 2e-12
Hs4505661 70.5 2e-12
CE02038 70.1 3e-12
CE28641 69.7 3e-12
Hs9506959 68.9 6e-12
Hs5453862 68.6 6e-12
Hs4505665 68.6 8e-12
7297876 68.2 1e-11
Hs20070127 65.9 4e-11
CE16340 63.9 2e-10
7290408 63.5 2e-10
Hs20538967 62.0 6e-10
CE23151 57.4 1e-08
Hs4505675 52.8 4e-07
CE24876 51.6 8e-07
Hs22051993 51.6 1e-06
7291615 48.5 8e-06
Hs19923440 44.7 1e-04
CE01047 44.3 1e-04
Hs15812216 41.2 0.001
Hs15812212 41.2 0.001
Hs15812214 40.8 0.001
Hs4505667 40.8 0.002
Hs5729972 40.0 0.003
Hs4505657 39.7 0.003
HsM5453866 39.7 0.004
Hs21361308 39.3 0.004
7298454 39.3 0.004
Hs4505669 34.3 0.15
Hs4585864 30.8 1.7
At3g43590 28.9 5.2
7298628 28.9 5.3
> Hs21361262
Length=545
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ HY+W+ ALM EFF QG +E ELGL SP+CD +T V +SQIGF
Sbjct 361 ILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTMVAQSQIGF 420
Query 119 LKLICFPLFESLAQA 133
+ I P F L +
Sbjct 421 IDFIVEPTFSLLTDS 435
> HsM4826892
Length=535
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ HY+W+ ALM EFF QG +E ELGL SP+CD +T V +SQIGF
Sbjct 361 ILHAADISHPAKSWKLHYRWTMALMEEFFLQGDKEAELGLPFSPLCDRKSTMVAQSQIGF 420
Query 119 LKLICFPLFESLAQA 133
+ I P F L +
Sbjct 421 IDFIVEPTFSLLTDS 435
> Hs4826894
Length=634
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ AD+ H A W+ H++W+ +L+ EFF QG +E ELGL SP+CD +T V +SQ+GF
Sbjct 371 MLHTADISHPAKAWDLHHRWTMSLLEEFFRQGDREAELGLPFSPLCDRKSTMVAQSQVGF 430
Query 119 LKLICFPLFESL 130
+ I P F L
Sbjct 431 IDFIVEPTFTVL 442
> Hs4505677
Length=536
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ AD+ H +W H +W++ALM EFF QG +E ELGL SP+CD +T V +SQIGF+
Sbjct 366 LHAADISHPTKQWLVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRTSTLVAQSQIGFI 425
Query 120 KLICFPLFESLA 131
I P F L
Sbjct 426 DFIVEPTFSVLT 437
> Hs4505663
Length=564
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 8/111 (7%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 378 TDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEISPMCDKHTA 437
Query 110 DVPKSQIGFLKLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
V KSQ+GF+ I PL+E+ L Q D A L L+DN +Q +
Sbjct 438 SVEKSQVGFIDYIVHPLWETWADLVQPD-----AQDILDTLEDNRNWYQSM 483
> Hs22050327
Length=641
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 60/106 (56%), Gaps = 2/106 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + + + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 438 SDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNA 497
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ A + A L L+DN E +Q
Sbjct 498 SVEKSQVGFIDYIVHPLWETWADLVHPD--AQDILDTLEDNREWYQ 541
> Hs5453864
Length=809
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 60/106 (56%), Gaps = 2/106 (1%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + + + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 606 SDRIQVLQNMVHCADLSNPTKPLQLYRQWTDRIMEEFFRQGDRERERGMEISPMCDKHNA 665
Query 110 DVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
V KSQ+GF+ I PL+E+ AD A L L+DN E +Q
Sbjct 666 SVEKSQVGFIDYIVHPLWETW--ADLVHPDAQDILDTLEDNREWYQ 709
> Hs4505661
Length=1112
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 52/175 (29%), Positives = 81/175 (46%), Gaps = 24/175 (13%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDFLA+F N+K N + I++ ++ D + + CI
Sbjct 894 DLKKHFDFLAEF-------NAKANDVNSNGIEW-------------SNENDRLLVCQVCI 933
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A + H KW++ ++NEF+ QG +E LGL +SP D ++ + K Q F+
Sbjct 934 KLADINGPAKVRDLHLKWTEGIVNEFYEQGDEEANLGLPISPFMDRSSPQLAKLQESFIT 993
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDN-SETWQRISDSEMDWREISQINKCN 174
I PL S D +G L L +DN +E+ E+D + N N
Sbjct 994 HIVGPLCNSY---DAAGLLPGQWLEAEEDNDTESGDDEDGEELDTEDEEMENNLN 1045
> CE02038
Length=549
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 51 DCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATD 110
D + ++ I AD+ + E + +W+Q +M E++ QG +E ELGL++SP+CD
Sbjct 427 DKIQVLQSMIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGLEISPMCDRGNVT 486
Query 111 VPKSQIGFLKLICFPLFESLA 131
+ KSQ+GF+ I PL+E+ A
Sbjct 487 IEKSQVGFIDYIVHPLYETWA 507
> CE28641
Length=626
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 51 DCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATD 110
D + ++ I AD+ + E + +W+Q +M E++ QG +E ELGL++SP+CD
Sbjct 504 DKIQVLQSMIHLADLSNPTKPIELYQQWNQRIMEEYWRQGDKEKELGLEISPMCDRGNVT 563
Query 111 VPKSQIGFLKLICFPLFESLA 131
+ KSQ+GF+ I PL+E+ A
Sbjct 564 IEKSQVGFIDYIVHPLYETWA 584
> Hs9506959
Length=450
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 0/110 (0%)
Query 48 DPADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEA 107
D D + + ++CAD+ + WE +WS+ + EF+ QG E + L++SP+C++
Sbjct 307 DAQDRHFMLQIALKCADICNPCRIWEMSKQWSERVCEEFYRQGELEQKFELEISPLCNQQ 366
Query 108 ATDVPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRI 157
+P QIGF+ I PLF A + L+ L L N W+ +
Sbjct 367 KDSIPSIQIGFMSYIVEPLFREWAHFTGNSTLSENMLGHLAHNKAQWKSL 416
> Hs5453862
Length=647
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 50/82 (60%), Gaps = 0/82 (0%)
Query 50 ADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAAT 109
+D + + + CAD+ + E + +W+ +M EFF QG +E E G+++SP+CD+
Sbjct 338 SDRIQVLRNMVHCADLSNPTKPLELYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTA 397
Query 110 DVPKSQIGFLKLICFPLFESLA 131
V KSQ+GF+ I PL+E+ A
Sbjct 398 SVEKSQVGFIDYIVHPLWETWA 419
> Hs4505665
Length=712
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 75/155 (48%), Gaps = 19/155 (12%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S H + LA L + V ++K + + L + +D + + +
Sbjct 500 DMSKHMNLLAD-------LKTMVETKKVTSLGV----------LLLDNYSDRIQVLQNLV 542
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
CAD+ + + +W+ +M EFF QG +E E GL +SP+CD+ V KSQ+GF+
Sbjct 543 HCADLSNPTKPLPLYRQWTDRIMAEFFQQGDRERESGLDISPMCDKHTASVEKSQVGFID 602
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
I PL+E+ AD A L L+DN E +Q
Sbjct 603 YIAHPLWETW--ADLVHPDAQDLLDTLEDNREWYQ 635
> 7297876
Length=605
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/82 (40%), Positives = 45/82 (54%), Gaps = 0/82 (0%)
Query 44 TLKTDPADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPV 103
TL+ D + + C D+ H A +W H++W+ L+ EFF QG E ELGL SP+
Sbjct 368 TLQEATIDKQKVLSLVLHCCDISHPAKQWGVHHRWTMLLLEEFFRQGDLEKELGLPFSPL 427
Query 104 CDEAATDVPKSQIGFLKLICFP 125
CD T V +SQI F+ I P
Sbjct 428 CDRNNTLVAESQICFIDFIVEP 449
> Hs20070127
Length=1141
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 63/133 (47%), Gaps = 22/133 (16%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HFDF+A+F N KVN + ID+ T+ D + + CI
Sbjct 909 DLKKHFDFVAKF-------NGKVNDDVG--IDW-------------TNENDRLLVCQMCI 946
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ A E H +W+ ++NEF+ QG +E LGL +SP D +A + Q F+
Sbjct 947 KLADINGPAKCKELHLQWTDGIVNEFYEQGDEEASLGLPISPFMDRSAPQLANLQESFIS 1006
Query 121 LICFPLFESLAQA 133
I PL S A
Sbjct 1007 HIVGPLCNSYDSA 1019
> CE16340
Length=664
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ D+ H A W H +W++ ++ EFF QG E +GL SP+CD V SQIGF+
Sbjct 476 VHACDISHPAKPWNLHERWTEGVLEEFFRQGDLEASMGLPYSPLCDRHTVHVADSQIGFI 535
Query 120 KLICFP 125
I P
Sbjct 536 DFIVEP 541
> 7290408
Length=624
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 53/101 (52%), Gaps = 8/101 (7%)
Query 60 IRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFL 119
+ CAD+ + + +W LM EFF QG +E E G+ +SP+CD + KSQ+GF+
Sbjct 440 VHCADLSNPTKPLPLYKRWVALLMEEFFLQGDKERESGMDISPMCDRHNATIEKSQVGFI 499
Query 120 KLICFPLFES---LAQADNSGDLAAICLSFLKDNSETWQRI 157
I PL+E+ L D A L L++N + +Q +
Sbjct 500 DYIVHPLWETWADLVHPD-----AQDILDTLEENRDYYQSM 535
> Hs20538967
Length=418
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query 57 KACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQI 116
+ ++CAD+ + WE +WS+ + EFF QG E + L VSP+CD + QI
Sbjct 291 QMALKCADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHTESIANIQI 350
Query 117 GFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQ 155
GF+ + PLF A+ N+ L+ L + N +W+
Sbjct 351 GFMTYLVEPLFTEWARFSNT-RLSQTMLGHVGLNKASWK 388
> CE23151
Length=760
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 75/159 (47%), Gaps = 28/159 (17%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
D+S HF++LA+F ++H + V E++ + T+ +
Sbjct 616 DISRHFEYLAKFN-KMHV--TDVPEEQRD--------------------TNSLTICDMLV 652
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVS-PVCDEAATDVPKSQIGFL 119
+CAD+ + A +W +W+ ++ E+F Q R+E E GL V+ V D +VP +Q GF+
Sbjct 653 KCADISNPAREWGLCQRWAHRIVEEYFEQTREEKEKGLPVTMEVFDRNTCNVPITQCGFI 712
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRIS 158
+ F + + G+L+ L+ N E W+ ++
Sbjct 713 DMFAREAFATFTEFAKLGELS----DQLESNYEKWKVMT 747
> Hs4505675
Length=593
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 55 LAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKS 114
L I+C D+ + E W L+ E+F Q +E GL V+P D +
Sbjct 453 LKMILIKCCDISNEVRPMEVAEPWVDCLLEEYFMQSDREKSEGLPVAPFMDRDKVTKATA 512
Query 115 QIGFLKLICFPLFESLAQ 132
QIGF+K + P+FE++ +
Sbjct 513 QIGFIKFVLIPMFETVTK 530
> CE24876
Length=562
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 42/178 (23%), Positives = 65/178 (36%), Gaps = 31/178 (17%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL HF+ + F +E+ +EID + D + K I
Sbjct 272 DLKQHFEIIMTF------------TERLTEIDVQVE-------------TDRLLIGKLLI 306
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLK 120
+ AD+ + H +W+ + EF+ QG E GL ++P D V K Q F+
Sbjct 307 KMADINSPTKPYGLHRQWTDRICEEFYEQGDDERRRGLPITPYMDRGDAQVAKLQDSFIA 366
Query 121 LICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREISQINKCNKYRC 178
+ P LA A N L I D SE + + W+E ++ Y
Sbjct 367 HVVSP----LATAMNECGLLPILPGL--DTSELIINMEHNHRKWKEQIELENGGSYEA 418
> Hs22051993
Length=761
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/160 (28%), Positives = 76/160 (47%), Gaps = 15/160 (9%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
+++ HF+ + +F +NS +N +EI+ S NP K P + + + I
Sbjct 604 EMTKHFEHVNKF------VNS-INKPMAAEIE-GSDCECNP--AGKNFPENQILIKRMMI 653
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVS-PVCDEAATDVPKSQIGFL 119
+CADV + + +W+ + E+FAQ +E GL V PV D +PKSQI F+
Sbjct 654 KCADVANPCRPLDLCIEWAGRISEEYFAQTDEEKRQGLPVVMPVFDRNTCSIPKSQISFI 713
Query 120 KLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISD 159
+F++ D L A+ + L DN + W+ + D
Sbjct 714 DYFITDMFDAW---DAFAHLPAL-MQHLADNYKHWKTLDD 749
> 7291615
Length=1060
Score = 48.5 bits (114), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 58/123 (47%), Gaps = 20/123 (16%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
+++ HF+ LA+F S + P+ NP +TD + + I
Sbjct 901 EMTRHFEHLAKF---------------VSVFGGEEPRDHNP----QTDEETSILMRRMLI 941
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQ-VSPVCDEAATDVPKSQIGFL 119
+ ADV + A + +W++ + E+F Q +E + L V P+ D A +PKSQIGF+
Sbjct 942 KVADVSNPARPMQFCIEWARRIAEEYFMQTDEEKQRHLPIVMPMFDRATCSIPKSQIGFI 1001
Query 120 KLI 122
+ I
Sbjct 1002 EYI 1004
> Hs19923440
Length=934
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 45/82 (54%), Gaps = 6/82 (7%)
Query 55 LAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQG-RQELELGLQVSPVCDEAATD-VP 112
L AC D+G WE + ++ + +EFF QG R+ LEL L S + D D +P
Sbjct 811 LMTAC----DLGAVTKPWEISRQVAELVTSEFFEQGDRERLELKLTPSAIFDRNRKDELP 866
Query 113 KSQIGFLKLICFPLFESLAQAD 134
+ Q+ ++ IC PL+++L + +
Sbjct 867 RLQLEWIDSICMPLYQALVKVN 888
> CE01047
Length=918
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 39/79 (49%), Gaps = 0/79 (0%)
Query 59 CIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGF 118
+ +D+ A + + ++ + EFFAQG EL+LG++ + D VP QI F
Sbjct 630 VMTASDLSDQAKNFHNAKRIAENIYLEFFAQGDLELQLGVKPLEMMDRTNAYVPTVQIDF 689
Query 119 LKLICFPLFESLAQADNSG 137
L I P+F+ LA G
Sbjct 690 LFKIGVPVFQLLASVVPEG 708
> Hs15812216
Length=823
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 72 WEQHYKWSQALMNEFFAQG-RQELELGLQVSPVCD-EAATDVPKSQIGFLKLICFPLFES 129
W + ++ + EFF QG R+ EL ++ + + + E +P Q+GF+ IC L+E+
Sbjct 720 WPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEA 779
Query 130 LAQADNSGDLAAICLSFL---KDNSETWQRISDSE 161
L C L + N + WQ +++ +
Sbjct 780 LTHVSED------CFPLLDGCRKNRQKWQALAEQQ 808
> Hs15812212
Length=833
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 72 WEQHYKWSQALMNEFFAQG-RQELELGLQVSPVCD-EAATDVPKSQIGFLKLICFPLFES 129
W + ++ + EFF QG R+ EL ++ + + + E +P Q+GF+ IC L+E+
Sbjct 730 WPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEA 789
Query 130 LAQADNSGDLAAICLSFL---KDNSETWQRISDSE 161
L C L + N + WQ +++ +
Sbjct 790 LTHVSED------CFPLLDGCRKNRQKWQALAEQQ 818
> Hs15812214
Length=865
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 72 WEQHYKWSQALMNEFFAQG-RQELELGLQVSPVCD-EAATDVPKSQIGFLKLICFPLFES 129
W + ++ + EFF QG R+ EL ++ + + + E +P Q+GF+ IC L+E+
Sbjct 762 WPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEA 821
Query 130 LAQADNSGDLAAICLSFL---KDNSETWQRISDSE 161
L C L + N + WQ +++ +
Sbjct 822 LTHVSED------CFPLLDGCRKNRQKWQALAEQQ 850
> Hs4505667
Length=875
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 72 WEQHYKWSQALMNEFFAQG-RQELELGLQVSPVCD-EAATDVPKSQIGFLKLICFPLFES 129
W + ++ + EFF QG R+ EL ++ + + + E +P Q+GF+ IC L+E+
Sbjct 772 WPIQQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEA 831
Query 130 LAQADNSGDLAAICLSFL---KDNSETWQRISDSE 161
L C L + N + WQ +++ +
Sbjct 832 LTHVSED------CFPLLDGCRKNRQKWQALAEQQ 860
> Hs5729972
Length=779
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query 85 EFFAQGRQELELGLQVSPVCDEAATD-VPKSQIGFLKLICFPLFESLAQA-DNSGDLAAI 142
EF+A+G + +LG+Q P+ D D VP+ Q+GF + P + +L Q + L
Sbjct 685 EFWAEGDEMKKLGIQPIPMMDRDKKDEVPQGQLGFYNAVAIPCYTTLTQILPPTEPLLKA 744
Query 143 CLSFLKDNSETWQRISDSE 161
C +DN W+++ E
Sbjct 745 C----RDNLSQWEKVIRGE 759
> Hs4505657
Length=941
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/106 (20%), Positives = 46/106 (43%), Gaps = 13/106 (12%)
Query 64 DVGHSAVKWEQHYKWSQALMNEFFAQGRQELELGLQVSPVCDEAATDVPKSQIGFLKLIC 123
D+ W+ K ++ + EFF+QG E +G + + D +P+ QI F++ I
Sbjct 808 DLSDQTKGWKTTRKIAELIYKEFFSQGDLEKAMGNRPMEMMDREKAYIPELQISFMEHIA 867
Query 124 FPLFESLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREISQ 169
P+++ L +E ++R++ + W ++S
Sbjct 868 MPIYKLLQ-------------DLFPKAAELYERVASNREHWTKVSH 900
> HsM5453866
Length=858
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 5/119 (4%)
Query 43 KTLKTDPADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELE-LGLQVS 101
K + DP + + D+ WE + + + NEF+ QG E L Q
Sbjct 702 KYVTVDPTKKEIIMAMMMTACDLSAITKPWEVQSQVALMVANEFWEQGDLERTVLQQQPI 761
Query 102 PVCDEAATD-VPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISD 159
P+ D D +PK Q+GF+ +C +++ ++ LS L++N W+ ++D
Sbjct 762 PMMDRNKRDELPKLQVGFIDFVCTFVYKEFSRFHKE---ITPMLSGLQNNRVEWKSLAD 817
> Hs21361308
Length=858
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 5/119 (4%)
Query 43 KTLKTDPADCWTLAKACIRCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELE-LGLQVS 101
K + DP + + D+ WE + + + NEF+ QG E L Q
Sbjct 702 KYVTVDPTKKEIIMAMMMTACDLSAITKPWEVQSQVALMVANEFWEQGDLERTVLQQQPI 761
Query 102 PVCDEAATD-VPKSQIGFLKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISD 159
P+ D D +PK Q+GF+ +C +++ ++ LS L++N W+ ++D
Sbjct 762 PMMDRNKRDELPKLQVGFIDFVCTFVYKEFSRFHKE---ITPMLSGLQNNRVEWKSLAD 817
> 7298454
Length=1284
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/91 (26%), Positives = 44/91 (48%), Gaps = 7/91 (7%)
Query 72 WEQHYKWSQALMNEFFAQGRQELELGLQVSPV---CDEAATDVPKSQIGFLKLICFPLFE 128
WE + + + +EFF QG E + L ++P+ E ++P Q+ F+ IC P++E
Sbjct 852 WEIEKRVADLVSSEFFEQGDMEKQ-ELNITPIDIMNREKEDELPMMQVNFIDSICLPIYE 910
Query 129 SLAQADNSGDLAAICLSFLKDNSETWQRISD 159
+ A D + ++DN W ++D
Sbjct 911 AFATLS---DKLEPLVEGVRDNRGHWIDLAD 938
> Hs4505669
Length=854
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 38/161 (23%), Positives = 70/161 (43%), Gaps = 18/161 (11%)
Query 1 DLSTHFDFLAQFRLQLHFLNSKVNSEKKSEIDFKSPQTLNPHKTLKTDPADCWTLAKACI 60
DL+ +F A F+ + SK +KKS +++ S +T + + AC
Sbjct 668 DLALYFKKRAMFQKIVD--ESKNYQDKKSWVEYLSLETTRKEIVMAM-------MMTAC- 717
Query 61 RCADVGHSAVKWEQHYKWSQALMNEFFAQGRQELE-LGLQVSPVCDE-AATDVPKSQIGF 118
D+ WE K + + EF+ QG E L Q P+ D A ++PK Q+GF
Sbjct 718 ---DLSAITKPWEVQSKVALLVAAEFWEQGDLERTVLDQQPIPMMDRNKAAELPKLQVGF 774
Query 119 LKLICFPLFESLAQADNSGDLAAICLSFLKDNSETWQRISD 159
+ +C +++ ++ L++N + W+ ++D
Sbjct 775 IDFVCTFVYKEFSRFHEE---ILPMFDRLQNNRKEWKALAD 812
> Hs4585864
Length=860
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 27/111 (24%), Positives = 52/111 (46%), Gaps = 8/111 (7%)
Query 72 WEQHYKWSQALMNEFFAQGRQELELGLQVSPVC---DEAATDVPKSQIGFLKLICFPLFE 128
WE + + + EF+ QG E + LQ +P+ A ++PK Q+GF+ +C +++
Sbjct 728 WEVQSQVALLVAAEFWEQGDLERTV-LQQNPIPMMDRNKADELPKLQVGFIDFVCTFVYK 786
Query 129 SLAQADNSGDLAAICLSFLKDNSETWQRISDSEMDWREISQINKCNKYRCA 179
++ L + +N + W+ ++D E D + Q K K + A
Sbjct 787 EFSRFHEE---ITPMLDGITNNRKEWKALAD-EYDAKMKVQEEKKQKQQSA 833
> At3g43590
Length=551
Score = 28.9 bits (63), Expect = 5.2, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 48 DPADCWTLAKACIRCADVGHSAVKWEQHYKWS 79
+P + + A +C RC +GHS + +HY+ S
Sbjct 276 EPGNSLSWAVSCYRCGQLGHSGLACGRHYEES 307
> 7298628
Length=1817
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 15/25 (60%), Positives = 18/25 (72%), Gaps = 1/25 (4%)
Query 21 SKVNSEKKSEIDFKSPQTLNPHKTL 45
S+VNS +KS DF P+TLNP K L
Sbjct 1562 SEVNSRRKS-FDFTRPKTLNPPKRL 1585
Lambda K H
0.320 0.132 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2914594326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40