bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
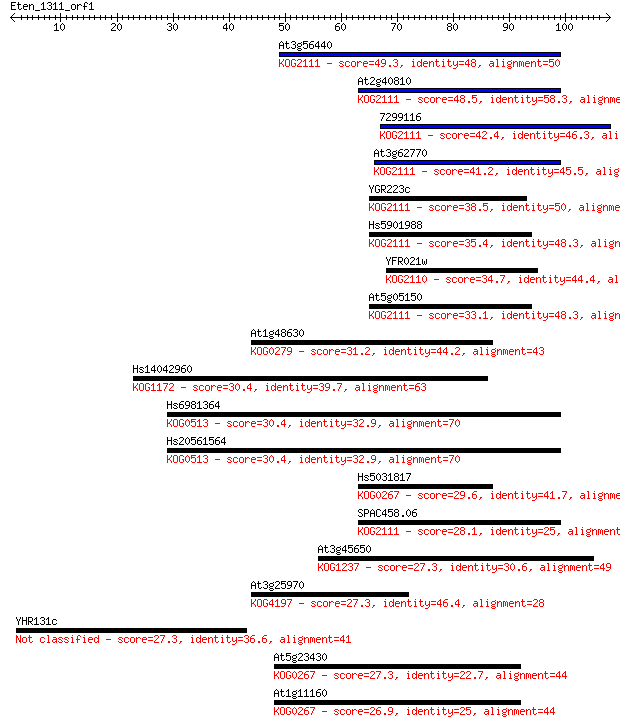
Query= Eten_1311_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
At3g56440 49.3 2e-06
At2g40810 48.5 3e-06
7299116 42.4 2e-04
At3g62770 41.2 5e-04
YGR223c 38.5 0.003
Hs5901988 35.4 0.025
YFR021w 34.7 0.048
At5g05150 33.1 0.13
At1g48630 31.2 0.43
Hs14042960 30.4 0.72
Hs6981364 30.4 0.75
Hs20561564 30.4 0.75
Hs5031817 29.6 1.4
SPAC458.06 28.1 3.6
At3g45650 27.3 6.3
At3g25970 27.3 6.3
YHR131c 27.3 7.2
At5g23430 27.3 7.5
At1g11160 26.9 9.1
> At3g56440
Length=400
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 49 TLGSVGIGNSASVYLESMAFNQDGSCFIAGTSEGFRVYTSSPLTEFVRRE 98
+ G G S L S+++NQD SCF AGTS GFR+Y P E RRE
Sbjct 22 SFGPPDFGESDEAELVSVSWNQDYSCFAAGTSHGFRIYNCEPFKETFRRE 71
> At2g40810
Length=369
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 63 LESMAFNQDGSCFIAGTSEGFRVYTSSPLTEFVRRE 98
L S+ +NQD SCF AGTS GFR+Y P E RRE
Sbjct 8 LVSVCWNQDSSCFAAGTSHGFRIYNCEPFKETFRRE 43
> 7299116
Length=340
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query 67 AFNQDGSCFIAGTSEGFRVYTSSPLTEFVRREGPWWPDRGV 107
AFNQD CF T GFRVY PL E +E ++P+ G+
Sbjct 17 AFNQDQGCFACATDTGFRVYNCDPLKE---KERQYFPEGGL 54
> At3g62770
Length=432
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 66 MAFNQDGSCFIAGTSEGFRVYTSSPLTEFVRRE 98
++FNQD +CF GT GFR+ P E RR+
Sbjct 81 LSFNQDHACFAVGTDRGFRILNCDPFREIFRRD 113
> YGR223c
Length=448
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 65 SMAFNQDGSCFIAGTSEGFRVYTSSPLT 92
S++FNQD SCF GFR++ + PLT
Sbjct 21 SVSFNQDDSCFSVALENGFRIFNTDPLT 48
> Hs5901988
Length=360
Score = 35.4 bits (80), Expect = 0.025, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 65 SMAFNQDGSCFIAGTSEGFRVYTSSPLTE 93
S+ FNQD SCF G R+Y PL E
Sbjct 11 SLRFNQDQSCFCCAMETGVRIYNVEPLME 39
> YFR021w
Length=500
Score = 34.7 bits (78), Expect = 0.048, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 68 FNQDGSCFIAGTSEGFRVYTSSPLTEF 94
FNQ G+C GTS+GF+++ P +F
Sbjct 13 FNQTGTCISLGTSKGFKIFNCEPFGKF 39
> At5g05150
Length=374
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 65 SMAFNQDGSCFIAGTSEGFRVYTSSPLTE 93
S+A+NQ S FI GT+ GF VY+ P+ +
Sbjct 35 SVAWNQVCSGFIVGTNHGFNVYSCKPMIK 63
> At1g48630
Length=326
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 44 AEAADTLGSVGIGNSASV-YLESMAFNQDGSCFIAGTSEG-FRVY 86
AEA T GS GIGN V Y S+ ++ DG+ +G ++G RV+
Sbjct 277 AEAEKTDGSTGIGNKTKVIYCTSLNWSADGNTLFSGYTDGVIRVW 321
> Hs14042960
Length=891
Score = 30.4 bits (67), Expect = 0.72, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 34/79 (43%), Gaps = 16/79 (20%)
Query 23 RGDARTDKAEGT--GSGDLSAGAAEAADTLGSVGIGNSASVY--------------LESM 66
RGD T + +G G+GDLSA AE + T+ G +S Y L
Sbjct 7 RGDRCTQEVQGLVHGAGDLSASLAENSPTMSQNGYFEDSSYYKCDTDDTFEAREEILGDE 66
Query 67 AFNQDGSCFIAGTSEGFRV 85
AF+ S ++G S F V
Sbjct 67 AFDTANSSIVSGESIRFFV 85
> Hs6981364
Length=806
Score = 30.4 bits (67), Expect = 0.75, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 7/72 (9%)
Query 29 DKAEGTGSGDL-SAGAAEAADTLGSVGIGNSASV-YLESMAFNQDGSCFIAGTSEGFRVY 86
+KA G + DL A + + ++ I +S S+ Y+ M F F G R Y
Sbjct 501 EKASGVATKDLFDWVAGTSTGGILALAILHSKSMAYMRGMYFRMKDEVF-----RGSRPY 555
Query 87 TSSPLTEFVRRE 98
S PL EF++RE
Sbjct 556 ESGPLEEFLKRE 567
> Hs20561564
Length=806
Score = 30.4 bits (67), Expect = 0.75, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 7/72 (9%)
Query 29 DKAEGTGSGDL-SAGAAEAADTLGSVGIGNSASV-YLESMAFNQDGSCFIAGTSEGFRVY 86
+KA G + DL A + + ++ I +S S+ Y+ M F F G R Y
Sbjct 501 EKASGVATKDLFDWVAGTSTGGILALAILHSKSMAYMRGMYFRMKDEVF-----RGSRPY 555
Query 87 TSSPLTEFVRRE 98
S PL EF++RE
Sbjct 556 ESGPLEEFLKRE 567
> Hs5031817
Length=655
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 63 LESMAFNQDGSCFIAGTSEGFRVY 86
+ S+ FN DG C +G + RVY
Sbjct 234 VRSVLFNPDGCCLYSGCQDSLRVY 257
> SPAC458.06
Length=364
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 9/36 (25%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 63 LESMAFNQDGSCFIAGTSEGFRVYTSSPLTEFVRRE 98
+ +++ NQD SC G++++ +PL +R+
Sbjct 4 INTVSLNQDASCMSVALDTGYKIFQINPLKLRAQRQ 39
> At3g45650
Length=558
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 21/49 (42%), Gaps = 0/49 (0%)
Query 56 GNSASVYLESMAFNQDGSCFIAGTSEGFRVYTSSPLTEFVRREGPWWPD 104
GN A Y E + + I G YTS+ L + ++R W PD
Sbjct 460 GNVALCYQEFPESMRSTATSITSVVIGICFYTSTALIDLIQRTTAWLPD 508
> At3g25970
Length=646
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 15/28 (53%), Gaps = 0/28 (0%)
Query 44 AEAADTLGSVGIGNSASVYLESMAFNQD 71
A A D LG G+ N A +ESM N D
Sbjct 495 AAAVDLLGRAGLVNKAKELIESMPLNPD 522
> YHR131c
Length=840
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 2 DIVQLSRQLLVTGMENWPQDDRGDARTDKAEGTGSGDLSAG 42
D+ Q SR L +T + DD TD+ E G D AG
Sbjct 599 DLSQSSRSLCLTNRDAEINDDESATETDEDENDGETDEYAG 639
> At5g23430
Length=922
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 10/44 (22%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 48 DTLGSVGIGNSASVYLESMAFNQDGSCFIAGTSEGFRVYTSSPL 91
+T +G G + + ++FN DG + G E ++++ P+
Sbjct 215 ETFELIGSGGPETAGVRCLSFNPDGKTVLCGLQESLKIFSWEPI 258
> At1g11160
Length=974
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 11/44 (25%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 48 DTLGSVGIGNSASVYLESMAFNQDGSCFIAGTSEGFRVYTSSPL 91
+T +G + + ++AF+ DG G +G +VY+ P+
Sbjct 163 ETFELIGTTRPEATGVRAIAFHPDGQTLFCGLDDGLKVYSWEPV 206
Lambda K H
0.313 0.132 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40