bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1307_orf2
Length=202
Score E
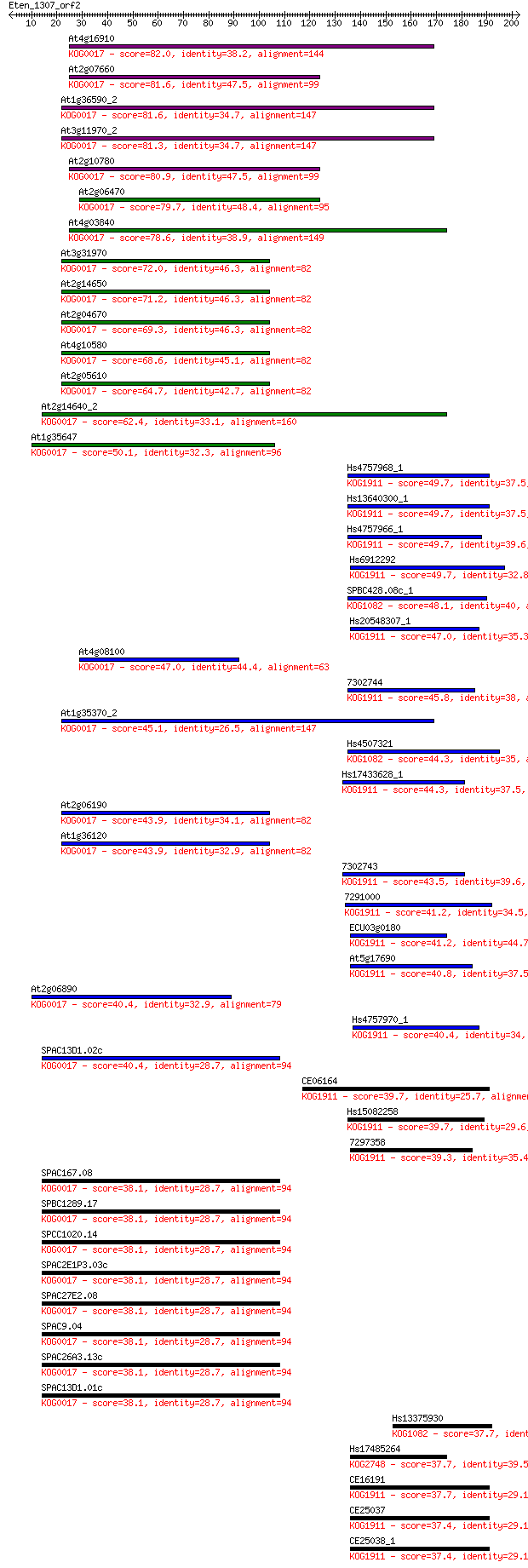
Sequences producing significant alignments: (Bits) Value
At4g16910 82.0 7e-16
At2g07660 81.6 1e-15
At1g36590_2 81.6 1e-15
At3g11970_2 81.3 1e-15
At2g10780 80.9 2e-15
At2g06470 79.7 3e-15
At4g03840 78.6 9e-15
At3g31970 72.0 8e-13
At2g14650 71.2 1e-12
At2g04670 69.3 5e-12
At4g10580 68.6 7e-12
At2g05610 64.7 1e-10
At2g14640_2 62.4 5e-10
At1g35647 50.1 3e-06
Hs4757968_1 49.7 4e-06
Hs13640300_1 49.7 4e-06
Hs4757966_1 49.7 4e-06
Hs6912292 49.7 4e-06
SPBC428.08c_1 48.1 1e-05
Hs20548307_1 47.0 2e-05
At4g08100 47.0 3e-05
7302744 45.8 6e-05
At1g35370_2 45.1 1e-04
Hs4507321 44.3 2e-04
Hs17433628_1 44.3 2e-04
At2g06190 43.9 2e-04
At1g36120 43.9 2e-04
7302743 43.5 3e-04
7291000 41.2 0.002
ECU03g0180 41.2 0.002
At5g17690 40.8 0.002
At2g06890 40.4 0.002
Hs4757970_1 40.4 0.002
SPAC13D1.02c 40.4 0.003
CE06164 39.7 0.004
Hs15082258 39.7 0.004
7297358 39.3 0.006
SPAC167.08 38.1 0.011
SPBC1289.17 38.1 0.013
SPCC1020.14 38.1 0.013
SPAC2E1P3.03c 38.1 0.013
SPAC27E2.08 38.1 0.013
SPAC9.04 38.1 0.013
SPAC26A3.13c 38.1 0.013
SPAC13D1.01c 38.1 0.013
Hs13375930 37.7 0.015
Hs17485264 37.7 0.016
CE16191 37.7 0.017
CE25037 37.4 0.019
CE25038_1 37.4 0.019
> At4g16910
Length=687
Score = 82.0 bits (201), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 55/157 (35%), Positives = 77/157 (49%), Gaps = 25/157 (15%)
Query 25 KAKWQQKYYADTKRRAVEYAMGDKVWLSS---KHLPPFTSCPKFEPRYRGPFEVIERIGT 81
+A+ +QK YA+ +R+ +E+ +GD V+L + K FTS K PRY GP++VIER+G
Sbjct 509 EAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGA 568
Query 82 VAYRLALPPTYEC-HDVFHVSQL--------VPHRPRPPALVSSAADAAWPP-IHDAAGN 131
VAY+L LPP + H+VFHVSQL PP L + AWP I D
Sbjct 569 VAYKLDLPPKLDAFHNVFHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKK 628
Query 132 PTAEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWE 168
T +D + + W + TWE
Sbjct 629 GTRGKSMDLLK------------ILWNCGGREEYTWE 653
> At2g07660
Length=949
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 47/111 (42%), Positives = 65/111 (58%), Gaps = 12/111 (10%)
Query 25 KAKWQQKYYADTKRRAVEYAMGDKVWLSS---KHLPPFTSCPKFEPRYRGPFEVIERIGT 81
+A+ +QK YA+ +R+ +E+ +GD V+L + K FTS K PRY GP++VIER+G
Sbjct 817 EAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGA 876
Query 82 VAYRLALPPTYEC-HDVFHVSQLVPH--------RPRPPALVSSAADAAWP 123
VAY+L LPP H+VFHVSQL + PP L + AWP
Sbjct 877 VAYKLDLPPKLNVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWP 927
> At1g36590_2
Length=958
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 51/155 (32%), Positives = 76/155 (49%), Gaps = 18/155 (11%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFT-------SCPKFEPRYRGPFE 74
H+++A+ + K +AD R E+ +GD V++ L P+ + K P+Y GP++
Sbjct 800 HLMRAQHRMKQFADQHRTEREFEIGDYVYVK---LQPYRQQSVVMRANQKLSPKYFGPYK 856
Query 75 VIERIGTVAYRLALPPTYECHDVFHVSQLVPHRPRPPALVSSAADAA-WPPIHDAAGNPT 133
+I+R G VAY+LALP + H VFHVSQL LV + + P +
Sbjct 857 IIDRCGEVAYKLALPSYSQVHPVFHVSQL-------KVLVGNVSTTVHLPSVMQDVFEKV 909
Query 134 AEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWE 168
E V+ M R + LVKW P + ATWE
Sbjct 910 PEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWE 944
> At3g11970_2
Length=958
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 51/155 (32%), Positives = 76/155 (49%), Gaps = 18/155 (11%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFT-------SCPKFEPRYRGPFE 74
H+++A+ + K +AD R E+ +GD V++ L P+ + K P+Y GP++
Sbjct 800 HLMRAQHRMKQFADQHRTEREFEIGDYVYVK---LQPYRQQSVVMRANQKLSPKYFGPYK 856
Query 75 VIERIGTVAYRLALPPTYECHDVFHVSQLVPHRPRPPALVSSAADAA-WPPIHDAAGNPT 133
+I+R G VAY+LALP + H VFHVSQL LV + + P +
Sbjct 857 IIDRCGEVAYKLALPSYSQVHPVFHVSQL-------KVLVGNVSTTVHLPSVMQDVFEKV 909
Query 134 AEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWE 168
E V+ M R + LVKW P + ATWE
Sbjct 910 PEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWE 944
> At2g10780
Length=1611
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 47/111 (42%), Positives = 64/111 (57%), Gaps = 12/111 (10%)
Query 25 KAKWQQKYYADTKRRAVEYAMGDKVWLSS---KHLPPFTSCPKFEPRYRGPFEVIERIGT 81
+A+ +QK YA+ +R+ +E+ +GD V+L + K FTS K PRY GP++VIER+G
Sbjct 1425 EAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGA 1484
Query 82 VAYRLALPPTYEC-HDVFHVSQL--------VPHRPRPPALVSSAADAAWP 123
VAY+L LPP H+VFHVSQL PP L + AWP
Sbjct 1485 VAYKLDLPPKLNAFHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWP 1535
> At2g06470
Length=899
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 46/107 (42%), Positives = 61/107 (57%), Gaps = 12/107 (11%)
Query 29 QQKYYADTKRRAVEYAMGDKVWLSS---KHLPPFTSCPKFEPRYRGPFEVIERIGTVAYR 85
+QK YA+ +R+ +E+ +GD V+L + K FTS K PRY GP++VIER+G VAY+
Sbjct 788 RQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 847
Query 86 LALPPTYEC-HDVFHVSQL--------VPHRPRPPALVSSAADAAWP 123
L LPP H+VFHVSQL PP L + AWP
Sbjct 848 LDLPPKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWP 894
> At4g03840
Length=973
Score = 78.6 bits (192), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 58/163 (35%), Positives = 79/163 (48%), Gaps = 27/163 (16%)
Query 25 KAKWQQKYYADTKRRAVEYAMGDKVWLSS---KHLPPFTSCPKFEPRYRGPFEVIERIGT 81
+A+ +QK YA+ +R+ +E+ + D V+L + K FTS K PRY GP++VIER+G
Sbjct 787 EAQDRQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGA 846
Query 82 VAYRLALPPTYEC-HDVFHVSQL--------VPHRPRPPALVSSAADAAWP-PIHDAAGN 131
VAY+L LPP H+VFHVSQL PP L + AWP I D
Sbjct 847 VAYKLDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTK 906
Query 132 PTAEYEVDYIMGQRG-SGDAAQYLVKWRGAPEDHATWEPAHHL 173
G RG S D + L W + TWE + +
Sbjct 907 -----------GTRGKSRDLLKVL--WNCGGREQYTWETENKM 936
> At3g31970
Length=1329
Score = 72.0 bits (175), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 53/86 (61%), Gaps = 4/86 (4%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHL---PPFTSCPKFEPRYRGPFEVIER 78
++ +A+ +Q+ YAD +RR +E+ +GD+V+L L S K PRY GPF ++ER
Sbjct 1163 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRYMGPFRIVER 1222
Query 79 IGTVAYRLALPPTYEC-HDVFHVSQL 103
+G VAYRL LP H VFHVS L
Sbjct 1223 VGPVAYRLELPDVMRAFHKVFHVSML 1248
> At2g14650
Length=1328
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 53/86 (61%), Gaps = 4/86 (4%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHL---PPFTSCPKFEPRYRGPFEVIER 78
++ +A+ +Q+ YAD +RR +E+ +GD+V+L L S K PRY GPF ++ER
Sbjct 1165 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVER 1224
Query 79 IGTVAYRLALPPTYEC-HDVFHVSQL 103
+G VAYRL LP H VFHVS L
Sbjct 1225 VGPVAYRLELPDVMRAFHKVFHVSML 1250
> At2g04670
Length=1411
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 53/86 (61%), Gaps = 4/86 (4%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHL-PPFTSC--PKFEPRYRGPFEVIER 78
++ +A+ +Q+ YAD +RR +E+ +GD+V+L L P S K PRY GPF ++ER
Sbjct 1245 NMKEAQARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVER 1304
Query 79 IGTVAYRLALPPTYEC-HDVFHVSQL 103
+G VAYRL LP H VFHV L
Sbjct 1305 VGPVAYRLELPDVMRAFHKVFHVLML 1330
> At4g10580
Length=1240
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/86 (43%), Positives = 53/86 (61%), Gaps = 4/86 (4%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHL---PPFTSCPKFEPRYRGPFEVIER 78
++ +A+ +Q+ YAD +RR +E+ +GD+V+L L S K PRY GPF+++ER
Sbjct 1074 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFKIVER 1133
Query 79 IGTVAYRLALPPTYEC-HDVFHVSQL 103
+ VAYRL LP H VFHVS L
Sbjct 1134 VEPVAYRLELPDVMRAFHKVFHVSML 1159
> At2g05610
Length=780
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 35/89 (39%), Positives = 51/89 (57%), Gaps = 10/89 (11%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFT-------SCPKFEPRYRGPFE 74
H+++A+ + K AD E+ +GD V++ L P+ S K P+Y GP++
Sbjct 669 HLMRAQHRMKQLADQHITEREFEVGDYVFVK---LQPYRQQSVVMRSTQKLSPKYFGPYK 725
Query 75 VIERIGTVAYRLALPPTYECHDVFHVSQL 103
VI+R G VAY+L LP + H VFHVSQL
Sbjct 726 VIDRCGEVAYKLQLPANSQVHPVFHVSQL 754
> At2g14640_2
Length=492
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 53/167 (31%), Positives = 72/167 (43%), Gaps = 14/167 (8%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFT----SCPKFEPRY 69
+L + H++ A K AD++ R V + +GD V L + T S K R+
Sbjct 316 ELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQPYRQKTLFRRSSQKLSHRF 375
Query 70 RGPFEVIERIGTVAYRLALPPTYECHDVFHVSQLVPHRPRPPALVSSAADAAWPPIHDAA 129
GPF+V + G VAYRL LP H VFHVS L +P PP+ +
Sbjct 376 YGPFQVASKHGEVAYRLTLPEGTRIHPVFHVSLL-----KPWVGDGEPDMGQLPPLRNNG 430
Query 130 G---NPTAEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHL 173
PTA EV + + A LV+W G + ATWE L
Sbjct 431 ELKLQPTAVLEVRW--RSQDKKRVADLLVQWEGLHIEDATWEEYDQL 475
> At1g35647
Length=1495
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 50/97 (51%), Gaps = 1/97 (1%)
Query 10 KLFRQLCDRAQSHILKAKWQQKYYADTKRRAVEYAMGDKVWLS-SKHLPPFTSCPKFEPR 68
+L +Q+ ++A+ +I + Q +A+ R+ V + GD VW+ K P K R
Sbjct 1243 ELVQQIHEQAKKNIEEKTKQYAKHANKSRKEVIFNEGDLVWIHLRKERFPKERKSKLMSR 1302
Query 69 YRGPFEVIERIGTVAYRLALPPTYECHDVFHVSQLVP 105
GPF+V++RI AY L L Y + F+V+ L P
Sbjct 1303 IDGPFKVLKRINNNAYSLDLQGKYNVSNSFNVADLFP 1339
> Hs4757968_1
Length=227
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 135 EYEVDYIMGQRGSGDA-AQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRR 190
E+EV+ I+ +R + QYLV+W+G + TWEP HL C + + RRQ +
Sbjct 5 EFEVEAIVDKRQDKNGNTQYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQTEK 61
> Hs13640300_1
Length=227
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 135 EYEVDYIMGQRGSGDA-AQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRR 190
E+EV+ I+ +R + QYLV+W+G + TWEP HL C + + RRQ +
Sbjct 5 EFEVEAIVDKRQDKNGNTQYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQTEK 61
> Hs4757966_1
Length=226
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 31/54 (57%), Gaps = 1/54 (1%)
Query 135 EYEVDYIMGQRGSGDA-AQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQ 187
E+EV+ I+ +R + QYLV+W+G + TWEP HL C + + RRQ
Sbjct 5 EFEVEAIVDKRQDKNGNTQYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQ 58
> Hs6912292
Length=191
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 36/61 (59%), Gaps = 1/61 (1%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRRLQARN 195
Y V+ ++ +R +YL+KW+G E+H TWEP +L CP L+ + ++ ++ + N
Sbjct 20 YVVEKVLDRRVVKGQVEYLLKWKGFSEEHNTWEPEKNL-DCPELISEFMKKYKKMKEGEN 78
Query 196 N 196
N
Sbjct 79 N 79
> SPBC428.08c_1
Length=126
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Query 135 EYEVDYIMGQR--GSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRR 189
EYEV+ I+ ++ +G Y ++W TWEP +L+GC A+L W+RR+RR
Sbjct 7 EYEVERIVDEKLDRNGAVKLYRIRWLNYSSRSDTWEPPENLSGCSAVLAEWKRRKRR 63
> Hs20548307_1
Length=227
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRR 186
+EV+ I+ + G Y V+W+G D TWEP HL C +L +R++
Sbjct 59 FEVEKILDMKTEGGKVLYKVRWKGYTSDDDTWEPEIHLEDCKEVLLEFRKK 109
> At4g08100
Length=1054
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Query 29 QQKYYADTKRRAVEYAMGDKVW--LSSKHLPPFTSCPKFEPRYRGPFEVIERIGTVAYRL 86
Q K YA+ R+ V + GD+VW L K P S K PR GPF+V++RI AY+L
Sbjct 905 QYKKYANKGRKEVIFNEGDQVWVHLRKKRFPEVRS-SKLMPRIDGPFKVLKRINNNAYKL 963
Query 87 ALPPT 91
L T
Sbjct 964 DLQDT 968
> 7302744
Length=418
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 135 EYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWR 184
EY V+ I+G+R Q LVKW G P ++ TWEP ++ C L+ +
Sbjct 23 EYVVEKILGKRFVNGRPQVLVKWSGFPNENNTWEPLENVGNCMKLVSDFE 72
> At1g35370_2
Length=923
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 63/154 (40%), Gaps = 39/154 (25%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFTS-------CPKFEPRYRGPFE 74
H+++A+ + K +AD R + +GD V++ L P+ K P+Y GP++
Sbjct 788 HLMRAQHRMKQFADQHRTERTFDIGDFVYVK---LQPYRQQSVVLRVNQKLSPKYFGPYK 844
Query 75 VIERIGTVAYRLALPPTYECHDVFHVSQLVPHRPRPPALVSSAADAAWPPIHDAAGNPTA 134
+IE+ G V V + P L P I + A
Sbjct 845 IIEKCGEVM----------------VGNVTTSTQLPSVL---------PDIFEKA----P 875
Query 135 EYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWE 168
EY ++ + +R A LVKW G P + ATW+
Sbjct 876 EYILERKLVKRQGRAATMVLVKWIGEPVEEATWK 909
> Hs4507321
Length=412
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 135 EYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRRLQAR 194
++EV+Y+ + + YLVKWRG P+ +TWEP +L C +L+ + + R L R
Sbjct 42 DFEVEYLCDYKKIREQEYYLVKWRGYPDSESTWEPRQNLK-CVRILKQFHKDLERELLRR 100
> Hs17433628_1
Length=179
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 133 TAEYEVDYIMGQR-GSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALL 180
+ E+EV+ I+ +R G +YLVKW+ + + TWEP HL C +
Sbjct 3 SQEFEVETIVDKRQGKNVNIEYLVKWKAYDKQYDTWEPKQHLMNCEKCM 51
> At2g06190
Length=280
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 44/88 (50%), Gaps = 6/88 (6%)
Query 22 HILKAKWQ-----QKYYADTKRRAVEYAMGDKVW-LSSKHLPPFTSCPKFEPRYRGPFEV 75
++KAK + K AD +RR + GD V L K + K +PR GPF+V
Sbjct 163 EVVKAKLEATGKKNKVAADKRRRFKVFKEGDDVMVLLRKGRFAVGTYNKVKPRKYGPFKV 222
Query 76 IERIGTVAYRLALPPTYECHDVFHVSQL 103
+ +I AY +ALP + + F+V+ +
Sbjct 223 LRKINDNAYVVALPKSMNISNTFNVADI 250
> At1g36120
Length=1235
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 41/83 (49%), Gaps = 27/83 (32%)
Query 22 HILKAKWQQKYYADTKRRAVEYAMGDKVWLSSKHLPPFTSCPKFEPRYRGPFEVIERIGT 81
++ +A+ +Q+ YAD +RR +E+ +G E++ER+G
Sbjct 1098 NMKEAQDRQRSYADKRRRELEFEVGT--------------------------EIVERVGP 1131
Query 82 VAYRLALPPTYEC-HDVFHVSQL 103
VAYRL LP H+VFHVS L
Sbjct 1132 VAYRLELPDVMRAFHNVFHVSML 1154
> 7302743
Length=84
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 133 TAEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALL 180
++EY V+ +G+R QYL KW G P + TWEP +L C L+
Sbjct 19 SSEYIVEKFLGKRYLRGRPQYLTKWEGYPIEQCTWEPLENLGKCMTLI 66
> 7291000
Length=240
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Query 134 AEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPAL-------LRTWRRR 186
AE+ V+ + +R +Y +KW+G P TWEP +L CP L L+ ++
Sbjct 2 AEFSVERVEDKRTVNGRTEYYLKWKGYPRSENTWEPVENL-DCPDLIANFEESLKNNKKE 60
Query 187 QRRRL 191
++RL
Sbjct 61 TKKRL 65
> ECU03g0180
Length=156
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHL 173
Y VD I+G R QYLVKW G P+ TWE ++
Sbjct 7 YTVDRIVGDRKKKGVKQYLVKWEGYPDSENTWEDEKNI 44
> At5g17690
Length=445
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTW 183
YE++ I +R QYL+KWRG PE TWEP +L ++ +
Sbjct 108 YEIEAIRRKRVRKGKVQYLIKWRGWPETANTWEPLENLQSIADVIDAF 155
> At2g06890
Length=1215
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 10 KLFRQLCDRAQSHILKAKWQQKYYADTKRRAVEYAMGDKVWLS-SKHLPPFTSCPKFEPR 68
+L +Q+ + A+ +I + A+ RR + +GD VW+ K P K PR
Sbjct 1022 ELVQQIHENARRNIEEKTKLYAKQANKGRREQIFEVGDMVWIHLRKERFPAQRKSKLMPR 1081
Query 69 YRGPFEVIERIGTVAYRLAL 88
GPF++I+RI AY+L L
Sbjct 1082 IDGPFKIIKRINDNAYQLDL 1101
> Hs4757970_1
Length=284
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 137 EVDYIMGQRGSGDA-AQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRR 186
+V+ I+ +R + +YLV+W+G + TWEP HL C + + RR
Sbjct 62 QVERIVDKRKNKKGKTEYLVRWKGYDSEDDTWEPEQHLVNCEEYIHDFNRR 112
> SPAC13D1.02c
Length=239
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 68 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 127
Query 73 FEVIERIGTVAYRLALPPTYE--CHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 128 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 164
> CE06164
Length=184
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 40/74 (54%), Gaps = 6/74 (8%)
Query 117 AADAAWPPIHDAAGNPTAEYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGC 176
A DA P +++ N + V+ ++ +R + ++Y +KW+G PE +WEP +L C
Sbjct 23 AQDA--PLFQESSSNV---FVVEKVLNKRLTRGGSEYYIKWQGFPESECSWEPIENLQ-C 76
Query 177 PALLRTWRRRQRRR 190
+++ + + +R
Sbjct 77 DRMIQEYEKEAAKR 90
> Hs15082258
Length=183
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 135 EYEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQR 188
E+ V+ ++ +R +Y +KW+G + TWEP +L CP L+ + Q+
Sbjct 29 EFVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEENLD-CPELIEAFLNSQK 81
> 7297358
Length=206
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTW 183
Y V+ I+ +R +Y +KW+G PE TWEP ++L C L++ +
Sbjct 24 YAVEKIIDRRVRKGKVEYYLKWKGYPETENTWEPENNLD-CQDLIQQY 70
> SPAC167.08
Length=1214
Score = 38.1 bits (87), Expect = 0.011, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1043 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1102
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1103 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1139
> SPBC1289.17
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPCC1020.14
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPAC2E1P3.03c
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPAC27E2.08
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPAC9.04
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPAC26A3.13c
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> SPAC13D1.01c
Length=1333
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 3/97 (3%)
Query 14 QLCDRAQSHILKAKWQQKYYADTKRRAVE-YAMGDKVWLSSKHLPPFTSCPKFEPRYRGP 72
Q+ + H+ + K Y D K + +E + GD V + K P + GP
Sbjct 1162 QVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTKTGFLHKSNKLAPSFAGP 1221
Query 73 FEVIERIGTVAYRLALPPT--YECHDVFHVSQLVPHR 107
F V+++ G Y L LP + + FHVS L +R
Sbjct 1222 FYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKYR 1258
> Hs13375930
Length=350
Score = 37.7 bits (86), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 153 YLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRRL 191
YLVKW+G P+ TWEP +L CP LL+ + + L
Sbjct 4 YLVKWKGWPDSTNTWEPLQNLK-CPLLLQQFSNDKHNYL 41
> Hs17485264
Length=259
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 136 YEVDYIMGQRGSGDAAQYLVKWRGAPEDHATWEPAHHL 173
+ V+ I +R +YLVKW+G P ++TWEP H+
Sbjct 11 FAVESIRKKRVRKGKVEYLVKWKGWPPKYSTWEPEEHI 48
> CE16191
Length=175
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 136 YEVDYIMGQR-GSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRR 190
+ V+ ++ +R G ++L++W+G PE ++WEP +L C +L + R +R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFEREFSKR 73
> CE25037
Length=301
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 136 YEVDYIMGQR-GSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRR 190
+ V+ ++ +R G ++L++W+G PE ++WEP +L C +L + R +R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFEREFSKR 73
> CE25038_1
Length=162
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 136 YEVDYIMGQR-GSGDAAQYLVKWRGAPEDHATWEPAHHLTGCPALLRTWRRRQRRR 190
+ V+ ++ +R G ++L++W+G PE ++WEP +L C +L + R +R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFEREFSKR 73
Lambda K H
0.321 0.135 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3551102874
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40