bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1246_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
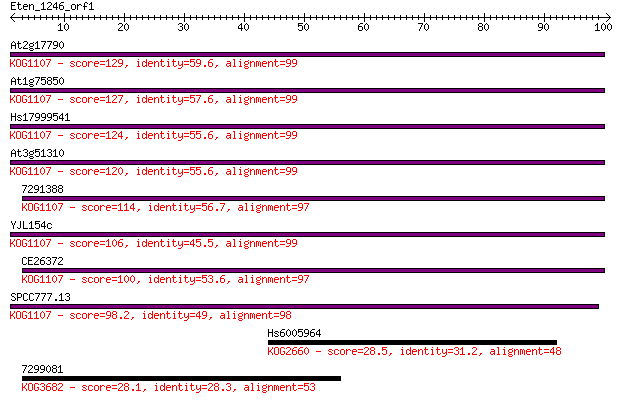
At2g17790 129 1e-30
At1g75850 127 3e-30
Hs17999541 124 4e-29
At3g51310 120 8e-28
7291388 114 3e-26
YJL154c 106 8e-24
CE26372 100 5e-22
SPCC777.13 98.2 3e-21
Hs6005964 28.5 3.4
7299081 28.1 4.3
> At2g17790
Length=830
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/99 (59%), Positives = 75/99 (75%), Gaps = 1/99 (1%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
GNI+PRLYLL T G +IK+KEAPA +IL+DL E+C+G+QHP+RGLFLR YL Q+ R+ L
Sbjct 99 GNILPRLYLLCTAGSVYIKTKEAPAKEILKDLVEMCRGIQHPLRGLFLRSYLAQISRDKL 158
Query 61 PDKGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
PD GSEYE + + DA F+L NF E +LWVR+Q Q
Sbjct 159 PDIGSEYEGDADTVI-DAVEFVLLNFTEMNKLWVRMQHQ 196
> At1g75850
Length=838
Score = 127 bits (320), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 57/99 (57%), Positives = 76/99 (76%), Gaps = 1/99 (1%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
GNI+PR+YLL TVG +IKSK+AP+ D+L+DL E+C+GVQHP+RGLFLR YL Q+ R+ L
Sbjct 128 GNILPRMYLLCTVGSVYIKSKQAPSKDVLKDLVEMCRGVQHPIRGLFLRSYLAQVSRDKL 187
Query 61 PDKGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
P+ GS+YE + + DA F+L NF E +LWVR+Q Q
Sbjct 188 PEIGSDYEGDANT-VMDAVEFVLQNFTEMNKLWVRIQHQ 225
> Hs17999541
Length=796
Score = 124 bits (311), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 55/99 (55%), Positives = 71/99 (71%), Gaps = 0/99 (0%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
GNI+PRLYLL+TVGV ++KS DIL+DL E+C+GVQHP+RGLFLR YL Q RN+L
Sbjct 102 GNIIPRLYLLITVGVVYVKSFPQSRKDILKDLVEMCRGVQHPLRGLFLRNYLLQCTRNIL 161
Query 61 PDKGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
PD+G ++E D+ F+L NF E +LWVR+Q Q
Sbjct 162 PDEGEPTDEETTGDISDSMDFVLLNFAEMNKLWVRMQHQ 200
> At3g51310
Length=789
Score = 120 bits (300), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 55/99 (55%), Positives = 72/99 (72%), Gaps = 1/99 (1%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
GNI+PRLYLL T+G +IKSK+ A DIL+DL E+C+ VQHP+RGLFLR YL Q+ R+ L
Sbjct 100 GNILPRLYLLCTIGSVYIKSKDVTATDILKDLVEMCRAVQHPLRGLFLRSYLAQVTRDKL 159
Query 61 PDKGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
P GS+ E +G + +A F+L NF E +LWVR+Q Q
Sbjct 160 PSIGSDLEGDGDAHM-NALEFVLQNFTEMNKLWVRMQHQ 197
> 7291388
Length=770
Score = 114 bits (286), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 55/97 (56%), Positives = 67/97 (69%), Gaps = 1/97 (1%)
Query 3 IVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVLPD 62
IVPRLYLL+TVG+ +IK+ IL+DL E+C+GVQHP+RGLFLR YL Q RN+LPD
Sbjct 97 IVPRLYLLITVGIVYIKNDPTLKRSILKDLVEMCRGVQHPLRGLFLRNYLLQCTRNILPD 156
Query 63 KGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
E E E DA F+LTNF E +LWVR+Q Q
Sbjct 157 VMVE-ENEHEGNVYDAIDFVLTNFAEMNKLWVRMQHQ 192
> YJL154c
Length=944
Score = 106 bits (265), Expect = 8e-24, Method: Composition-based stats.
Identities = 45/99 (45%), Positives = 68/99 (68%), Gaps = 10/99 (10%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
GN+VPRLYL++TVG +++ EAP +IL+D+ E+C+GVQ+P+RGLFLR+YL+Q + +L
Sbjct 93 GNVVPRLYLMITVGTSYLTFNEAPKKEILKDMIEMCRGVQNPIRGLFLRYYLSQRTKELL 152
Query 61 PDKGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
P+ + + F++ NF E +LWVRLQ Q
Sbjct 153 PEDDPSFNSQ----------FIMNNFIEMNKLWVRLQHQ 181
> CE26372
Length=793
Score = 100 bits (250), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 66/113 (58%), Gaps = 16/113 (14%)
Query 3 IVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVLPD 62
I+PRLYLLVT+G FIK +IL+DL E+C+GVQHP+RGLFLR YL Q R+VLPD
Sbjct 76 IIPRLYLLVTIGGVFIKCGLGSRKEILKDLVEMCRGVQHPLRGLFLRNYLMQCTRSVLPD 135
Query 63 ----------------KGSEYEQEGGSGAEDAFLFLLTNFNESARLWVRLQQQ 99
KG+ + +D F+L NF E +LWVR+Q Q
Sbjct 136 FPETEEMLVAHNDNLSKGTPKLKPRDGTVDDTIDFVLINFAEMNKLWVRMQHQ 188
> SPCC777.13
Length=785
Score = 98.2 bits (243), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 48/100 (48%), Positives = 67/100 (67%), Gaps = 3/100 (3%)
Query 1 GNIVPRLYLLVTVGVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQLCRNVL 60
G+IVPRLYL++TVG A++++ A +I+ DL ++C+GVQHP+RGLFLR YL R L
Sbjct 51 GSIVPRLYLMITVGSAYLETPNALVREIMNDLLDMCRGVQHPLRGLFLRHYLLTQTRKGL 110
Query 61 PDKGSEYEQEGGSGAE--DAFLFLLTNFNESARLWVRLQQ 98
P GSE E++ D+ FL+ NF E +LWVR+Q
Sbjct 111 P-LGSEDEEDASRKGTVLDSVKFLVINFTEMNKLWVRIQH 149
> Hs6005964
Length=344
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 44 RGLFLRFYLTQLCRNVLPDKGSEYEQEGGSGAEDAFLFLLTNFNESAR 91
R + + LT++ D+G EQE G+ ++D + L F E AR
Sbjct 95 RDFYAAYPLTEVPNGSNEDRGEVLEQEKGALSDDEIVSLSIEFYEGAR 142
> 7299081
Length=979
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query 3 IVPRLYLLVTV--GVAFIKSKEAPAADILRDLTELCKGVQHPMRGLFLRFYLTQL 55
++PR YL +++ F+ S IL+ LT +G+ P+ + R YL ++
Sbjct 274 LLPRFYLELSIFKCYEFLSSSREEYERILQRLTHQLRGIADPLVSSYARCYLVRM 328
Lambda K H
0.323 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40