bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1228_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
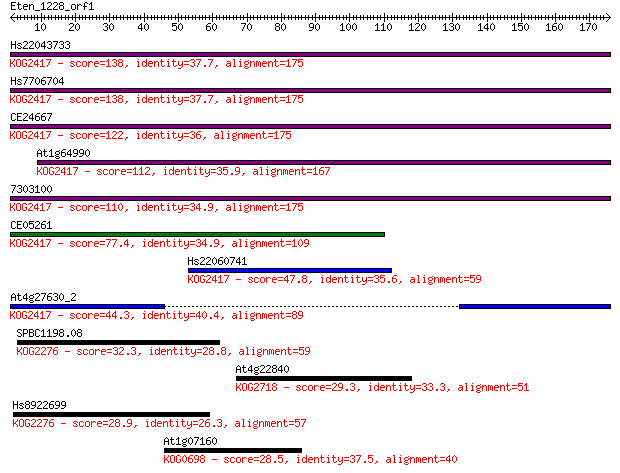
Hs22043733 138 6e-33
Hs7706704 138 6e-33
CE24667 122 3e-28
At1g64990 112 5e-25
7303100 110 1e-24
CE05261 77.4 1e-14
Hs22060741 47.8 1e-05
At4g27630_2 44.3 1e-04
SPBC1198.08 32.3 0.54
At4g22840 29.3 4.6
Hs8922699 28.9 5.0
At1g07160 28.5 6.5
> Hs22043733
Length=298
Score = 138 bits (348), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 66/175 (37%), Positives = 111/175 (63%), Gaps = 1/175 (0%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVL 60
E++ALEELSR+LF+ DL ++ + Y +T+ G+ N LG+ + CV+++FM+ N++
Sbjct 93 EVDALEELSRQLFLETADLYATKERIEYSKTFKGKYFNFLGYFFSIYCVWKIFMATINIV 152
Query 61 LDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTVFRY 120
DRV TDP TR +E+ ++ L + D+ + ++S +L+G II +IRG L F
Sbjct 153 FDRVGKTDPVTRGIEITVNYLGIQFDVKFWSQHISFILVGIIIVTSIRGLLITLTKFFYA 212
Query 121 MSTAVSSNVFALVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
+S++ SSNV L+++++MGMYF + +LL R+ +P YR + +VL L F +H
Sbjct 213 ISSSKSSNVIVLLLAQIMGMYFVSSVLLIRMSMPLEYRTIITEVLG-ELQFNFYH 266
> Hs7706704
Length=455
Score = 138 bits (347), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 66/175 (37%), Positives = 111/175 (63%), Gaps = 1/175 (0%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVL 60
E++ALEELSR+LF+ DL ++ + Y +T+ G+ N LG+ + CV+++FM+ N++
Sbjct 250 EVDALEELSRQLFLETADLYATKERIEYSKTFKGKYFNFLGYFFSIYCVWKIFMATINIV 309
Query 61 LDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTVFRY 120
DRV TDP TR +E+ ++ L + D+ + ++S +L+G II +IRG L F
Sbjct 310 FDRVGKTDPVTRGIEITVNYLGIQFDVKFWSQHISFILVGIIIVTSIRGLLITLTKFFYA 369
Query 121 MSTAVSSNVFALVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
+S++ SSNV L+++++MGMYF + +LL R+ +P YR + +VL L F +H
Sbjct 370 ISSSKSSNVIVLLLAQIMGMYFVSSVLLIRMSMPLEYRTIITEVLG-ELQFNFYH 423
> CE24667
Length=465
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 63/177 (35%), Positives = 108/177 (61%), Gaps = 3/177 (1%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVL 60
EI LE L+R LF+ L +L + + +T++G N+LG + C++++F+S N++
Sbjct 259 EIIPLETLARFLFLDLVELRQMLNRVEFSKTFMGIYFNILGHFFSIYCIWKIFISFINIV 318
Query 61 LDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTVFRY 120
DRV DP T+T+E+ +H + +P+DI+ + Y+S L+G I ++RG + F
Sbjct 319 FDRVGKVDPVTKTIEIGVHWMGIPLDISFWSQYISFFLVGVIAVTSVRGLLITMTKFFVS 378
Query 121 MSTAVS--SNVFALVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
+S A S SN+ AL+M+++MGMYF + +LL R+ +P+ YR L ++L L F +H
Sbjct 379 ISNATSSLSNIIALLMAQIMGMYFVSSVLLMRMNVPEEYRTILTRILG-DLKFNFYH 434
> At1g64990
Length=465
Score = 112 bits (279), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 60/170 (35%), Positives = 96/170 (56%), Gaps = 4/170 (2%)
Query 9 SRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVLLDRVSTTD 68
S++LF+ + +L ++ A Y RTW G + N+LG+ + CVY++ S +V+ T D
Sbjct 264 SKQLFLEVYELRQAKDAAAYSRTWKGHVQNLLGYACSIYCVYKMLKSLQSVVFKEAGTKD 323
Query 69 PATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTVF---RYMSTAV 125
P T + + L L + VD LL+ Y+SL+ +G +I +++RGF L+ F + +
Sbjct 324 PVTTMISIFLRLFDIGVDAALLSQYISLLFIGMLIVISVRGFLTNLMKFFFAVSRVGSGS 383
Query 126 SSNVFALVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
SSNV L +SE+MGMYF + +LL R L YR + VL + F +H
Sbjct 384 SSNV-VLFLSEIMGMYFLSSILLIRKSLRNEYRGIITDVLGGDIQFDFYH 432
> 7303100
Length=441
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 61/175 (34%), Positives = 100/175 (57%), Gaps = 1/175 (0%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVL 60
E+ LEEL R +F+ L L + + + +T G+ N+LG + CVY++FM N++
Sbjct 235 EVYGLEELLRSVFLELSSLKNMEERQRWSQTLKGKYFNVLGHFFSVYCVYKIFMCCINII 294
Query 61 LDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTVFRY 120
DRV DP TR LE+ +H +D ++S +L+G I+ +IRG L F
Sbjct 295 FDRVGRKDPVTRGLEIAIHWCGFNIDFAFWNQHISFLLVGCIVITSIRGLLLTLTKFFYR 354
Query 121 MSTAVSSNVFALVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
+S++ SSN+ L++ ++MGMYF + +LL R+ +P YR + +VL +L F +H
Sbjct 355 ISSSKSSNIIVLILGQIMGMYFCSSVLLMRMNMPAEYRVIITEVLG-NLHFNFYH 408
> CE05261
Length=434
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 38/109 (34%), Positives = 67/109 (61%), Gaps = 0/109 (0%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMTAVCVYRVFMSAANVL 60
EI+ LE LSR LF+ L +L + + + +T++G N+LG + CV+++F+S N+L
Sbjct 256 EIKPLETLSRYLFLELVELRNMLERVAFSKTFIGIYFNVLGHFFSLYCVWKIFISLVNIL 315
Query 61 LDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRG 109
DRV DP TR +E+ ++ + + +D+ + Y+S L+G I ++RG
Sbjct 316 FDRVGKVDPVTRLIEVSVNYVGIDMDVRYWSQYISFFLVGVIAITSVRG 364
> Hs22060741
Length=559
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 0/59 (0%)
Query 53 FMSAANVLLDRVSTTDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFF 111
F N+ DRV DP TR +E+ ++ L + D+ + ++S +L+G IIA +IRG
Sbjct 237 FHGTINIAFDRVGKMDPVTRGVEITVNYLGIQCDVKFWSQHISSILVGIIIATSIRGLL 295
> At4g27630_2
Length=418
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 1 EIEALEELSRELFVGLDDLVHSRAQALYGRTWLGRLNNMLGWGMT 45
E+E LEELS++LF+ + +L ++ A + RTW G + N LG+ +
Sbjct 258 EVEGLEELSKQLFLEIYELRQAKDAAAFSRTWKGHVQNFLGYACS 302
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 132 LVMSEVMGMYFSACLLLTRVYLPQSYRDALAQVLAPSLDFRAFH 175
L +SE+MGMYF + +LL R L YR + VL + F +H
Sbjct 342 LFLSEIMGMYFLSSILLIRKSLRNEYRGIITDVLGGDIQFDFYH 385
> SPBC1198.08
Length=474
Score = 32.3 bits (72), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 31/68 (45%), Gaps = 12/68 (17%)
Query 3 EALEELSRELFVGLDDLVHSRAQALYGRT---------WLGRLNNMLGWGMTAVCVYRVF 53
E +EE E GL+DL+ + A+ + + WLG +L +G+ VC + +
Sbjct 162 EGMEEYGSE---GLEDLIRAEAEKYFAKADCVCISDTYWLGTKKPVLTYGLRGVCYFNIT 218
Query 54 MSAANVLL 61
+ + L
Sbjct 219 VEGPSADL 226
> At4g22840
Length=460
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 67 TDPATRTLELLLHLLHVPVDITLLTPYLSLVLLGWIIALTIRGFFEKLLTV 117
TDPA L +++ L + L+TP LSL+L+G + + ++G +L V
Sbjct 263 TDPALAPLSIVMTSLSTATAV-LVTPMLSLLLIGKKLPVDVKGMISSILQV 312
> Hs8922699
Length=475
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 15/66 (22%), Positives = 30/66 (45%), Gaps = 12/66 (18%)
Query 2 IEALEELSRELFVGLDDLVHSRAQALYGRT---------WLGRLNNMLGWGMTAVCVYRV 52
+E +EE E GLD+L+ +R + WLG+ + +G+ +C + +
Sbjct 162 LEGMEESGSE---GLDELIFARKDTFFKDVDYVCISDNYWLGKKKPCITYGLRGICYFFI 218
Query 53 FMSAAN 58
+ +N
Sbjct 219 EVECSN 224
> At1g07160
Length=380
Score = 28.5 bits (62), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 46 AVCVYRVFMSAANVLLDRVSTTDPATRTLELLLHLLHVPV 85
AVC VF ++++ ++ T PA TL L L L+ PV
Sbjct 6 AVCNSPVFSPSSSLFCNKPLNTSPAHETLTLSLSHLNPPV 45
Lambda K H
0.329 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40