bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1227_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
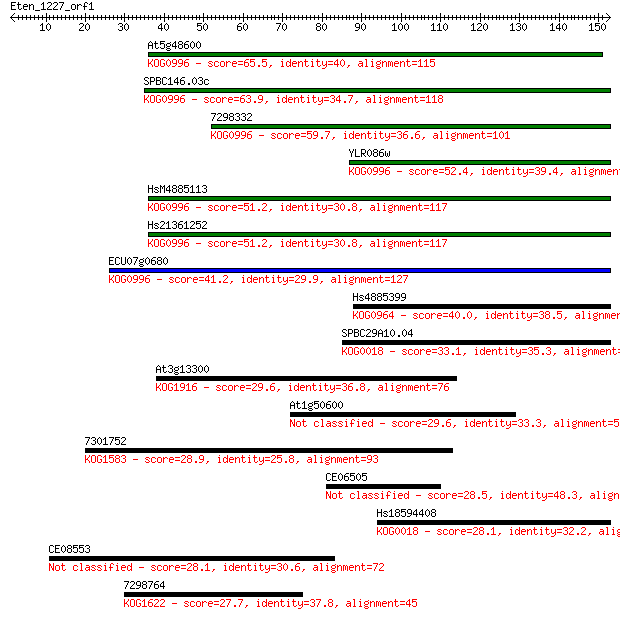
At5g48600 65.5 4e-11
SPBC146.03c 63.9 1e-10
7298332 59.7 2e-09
YLR086w 52.4 4e-07
HsM4885113 51.2 8e-07
Hs21361252 51.2 8e-07
ECU07g0680 41.2 9e-04
Hs4885399 40.0 0.002
SPBC29A10.04 33.1 0.23
At3g13300 29.6 2.1
At1g50600 29.6 2.6
7301752 28.9 3.7
CE06505 28.5 4.9
Hs18594408 28.1 6.3
CE08553 28.1 6.8
7298764 27.7 8.4
> At5g48600
Length=1241
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 46/120 (38%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSG--RLAEAEAEAARCTDTSAPR 93
Y VVE S A ELLR LG +LEK+ +L E T PR
Sbjct 584 YIVVETTSSAQACVELLRKGNLGFAT---FMILEKQTDHIHKLKEKVK-----TPEDVPR 635
Query 94 LLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALR---QRVVTLEGGLIELDGRM 150
L DL+ R K+AF+ ++GNT VA D++ AT +AY +RVV L+G L E G M
Sbjct 636 LFDLVRVKDERMKLAFYAALGNTVVAKDLDQATRIAYGGNREFRRVVALDGALFEKSGTM 695
> SPBC146.03c
Length=1324
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 41/119 (34%), Positives = 59/119 (49%), Gaps = 10/119 (8%)
Query 35 SYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRL 94
++ VV+ + LR LGR + + L+ L ++ R+ E + PRL
Sbjct 679 NHIVVDNIETGQKCVAFLRSNNLGRASFIILKELAQKNLARIQTPE---------NVPRL 729
Query 95 LDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA-LRQRVVTLEGGLIELDGRMVG 152
DLL F +F AF+ + NT VA ++E A +AY R RVVTL G LI+ G M G
Sbjct 730 FDLLRFNDQKFAPAFYNVLQNTLVAKNLEQANRIAYGKTRWRVVTLSGQLIDKSGTMTG 788
> 7298332
Length=1409
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 37/102 (36%), Positives = 52/102 (50%), Gaps = 7/102 (6%)
Query 52 LRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLLDLLHFISPRFKIAFFK 111
L+ Y +GR + L +E EA + T + PRL DL+ R + AF+
Sbjct 663 LKEYNVGRATFITLDKIEHH------RREANSRINTPENVPRLYDLVKVEDDRVRTAFYF 716
Query 112 SVGNTRVAADMEHATHLAYAL-RQRVVTLEGGLIELDGRMVG 152
++ NT V D+E T +AY R RVVTL G +IE+ G M G
Sbjct 717 ALRNTLVCDDLEQGTRIAYGRERYRVVTLRGEMIEMTGTMSG 758
> YLR086w
Length=1418
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query 87 TDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYALRQ-RVVTLEGGLIE 145
T + PRL DL+ +P+F AF+ + +T VA +++ A ++AY ++ RVVT++G LI+
Sbjct 757 TPENVPRLFDLVKPKNPKFSNAFYSVLRDTLVAQNLKQANNVAYGKKRFRVVTVDGKLID 816
Query 146 LDGRMVG 152
+ G M G
Sbjct 817 ISGTMSG 823
> HsM4885113
Length=1288
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 59/119 (49%), Gaps = 9/119 (7%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLL 95
Y VV+ A E L+ +G + L + + ++ E + T + PRL
Sbjct 642 YIVVDSIDIAQECVNFLKRQNIGVATFIGLDKMAV-WAKKMTEIQ------TPENTPRLF 694
Query 96 DLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLIELDGRMVG 152
DL+ + + AF+ ++ +T VA +++ AT +AY R RVVTL+G +IE G M G
Sbjct 695 DLVKVKDEKIRQAFYFALRDTLVADNLDQATRVAYQKDRRWRVVTLQGQIIEQSGTMTG 753
> Hs21361252
Length=1288
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 59/119 (49%), Gaps = 9/119 (7%)
Query 36 YYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAARCTDTSAPRLL 95
Y VV+ A E L+ +G + L + + ++ E + T + PRL
Sbjct 642 YIVVDSIDIAQECVNFLKRQNIGVATFIGLDKMAV-WAKKMTEIQ------TPENTPRLF 694
Query 96 DLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLIELDGRMVG 152
DL+ + + AF+ ++ +T VA +++ AT +AY R RVVTL+G +IE G M G
Sbjct 695 DLVKVKDEKIRQAFYFALRDTLVADNLDQATRVAYQKDRRWRVVTLQGQIIEQSGTMTG 753
> ECU07g0680
Length=1112
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 56/128 (43%), Gaps = 15/128 (11%)
Query 26 FLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSGRLAEAEAEAAR 85
F A G SS VV+ A E +++ LGR + L + +E
Sbjct 518 FRVAGKGLSSI-VVDTTCTAEECISVIKKLGLGRATFIILDRI------------SEVPV 564
Query 86 CTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA-LRQRVVTLEGGLI 144
S P + L+ F+ F+ ++ +T V +E A LA+ R+RVVTL+G LI
Sbjct 565 LPRESVPYMYSLIR-CGEEFRKCFYFALKDTLVCDGLEQAERLAFGKQRKRVVTLDGKLI 623
Query 145 ELDGRMVG 152
E G M G
Sbjct 624 EKSGVMSG 631
> Hs4885399
Length=1217
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query 88 DTSAPRLLDLLHFIS-----PRFKIAFFKSVGNTRVAADMEHATHLAYALRQRVVTLEGG 142
DT+ P D + IS PRF AF G T + ME +T LA A +TLEG
Sbjct 597 DTAYPETNDAIPMISKLRYNPRFDKAFKHVFGKTLICRSMEVSTQLARAFTMDCITLEGD 656
Query 143 LIELDGRMVG 152
+ G + G
Sbjct 657 QVSHRGALTG 666
> SPBC29A10.04
Length=1233
Score = 33.1 bits (74), Expect = 0.23, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 85 RCTDTSAPRLLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAY--ALRQRVVTLEGG 142
R T A +D+L+F S ++ +VGNT + M A L+Y L + VTLEG
Sbjct 593 RGTHKGARLAIDVLNFES-EYERVMISAVGNTLICDSMTVARDLSYNKRLNAKTVTLEGT 651
Query 143 LIELDGRMVG 152
+I G + G
Sbjct 652 VIHKTGLITG 661
> At3g13300
Length=1326
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 28/87 (32%), Positives = 37/87 (42%), Gaps = 16/87 (18%)
Query 38 VVEKPSDAHELFELLRVYELGRCNALALQV---LEKEL--------SGRLAEAEAEAARC 86
VV +P D H + V R + + L L +E+ G L A+AE+ RC
Sbjct 393 VVLRPHDGHPVSSATFVTSPERPDHIILITGGPLNREMKIWVSAGEEGWLLPADAESWRC 452
Query 87 TDTSAPRLLDLLHFISPRFKIAFFKSV 113
T T LDL PR + AFF V
Sbjct 453 TQT-----LDLKSSTEPRAEEAFFNQV 474
> At1g50600
Length=526
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 28/63 (44%), Gaps = 6/63 (9%)
Query 72 LSGRLAEAEA---EAARCTDTSAPRLLDLLHFI---SPRFKIAFFKSVGNTRVAADMEHA 125
L RLA + + +A RC D + P LL +H + P FK + + G A E
Sbjct 204 LVARLASSGSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESF 263
Query 126 THL 128
H+
Sbjct 264 VHI 266
> 7301752
Length=322
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 44/99 (44%), Gaps = 15/99 (15%)
Query 20 LLPLPLFLAASGGFSSYYVVEKPSDAHEL------FELLRVYELGRCNALALQVLEKELS 73
LLPLP FL +++++ ++++L L+ +Y LG VL + L
Sbjct 205 LLPLPAFLLMHDDIRTHWLLAFTGESYQLPLLGVAVPLILLYLLG-------NVLAQHLC 257
Query 74 GRLAEAEAEAARCTDTSAPRLLDLLHFISPRFKIAFFKS 112
++ C+ + +L L FIS F I +F++
Sbjct 258 --ISSVYTLTTECSSLTVTLILTLRKFISLVFSIVYFRN 294
> CE06505
Length=610
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 81 AEAARCTDTSAPRLLDLLHFISPRFKIAF 109
AE+A T+TSA LLD LH + + +AF
Sbjct 568 AESASATNTSATNLLDALHSSALKCGVAF 596
> Hs18594408
Length=1202
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 2/61 (3%)
Query 94 LLDLLHFISPRFKIAFFKSVGNTRVAADMEHATHLAYA--LRQRVVTLEGGLIELDGRMV 151
++D++ P+ K GN V ME A H+A + RQ+ V L+G L G +
Sbjct 593 VIDVIKTQFPQLKKVIQFVCGNGLVCETMEEARHIALSGPERQKTVALDGTLFLKSGVIS 652
Query 152 G 152
G
Sbjct 653 G 653
> CE08553
Length=461
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 0/72 (0%)
Query 11 PLPLSLLLLLLPLPLFLAASGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEK 70
P PL L L LP +G F S+Y P+ F +LR+ + NAL L +
Sbjct 113 PSPLCLKLFSLPFAPSAHLTGPFISHYPHYSPNMDAVSFPILRLPQPALKNALRQMFLVE 172
Query 71 ELSGRLAEAEAE 82
+S + A+A+
Sbjct 173 HISLSVLSAKAK 184
> 7298764
Length=683
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Query 30 SGGFSSYYVVEKPSDAHELFELLRVYELGRCNALALQVLEKELSG 74
SGG +S Y + PS +LF+ L++ LG C +Q++ KE G
Sbjct 66 SGGPNSVYAEDAPSYDPDLFK-LKIPMLGIC--YGMQLINKEFGG 107
Lambda K H
0.326 0.142 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40