bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
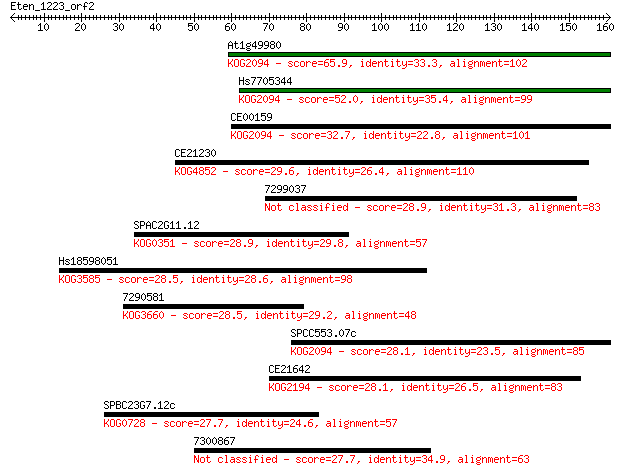
Query= Eten_1223_orf2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
At1g49980 65.9 3e-11
Hs7705344 52.0 5e-07
CE00159 32.7 0.27
CE21230 29.6 2.4
7299037 28.9 4.8
SPAC2G11.12 28.9 4.9
Hs18598051 28.5 5.4
7290581 28.5 5.9
SPCC553.07c 28.1 6.7
CE21642 28.1 7.8
SPBC23G7.12c 27.7 8.7
7300867 27.7 9.6
> At1g49980
Length=795
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 60/102 (58%), Gaps = 5/102 (4%)
Query 59 IFANSKAGMDMTEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRRVQLFASS 118
+F N+KAGM+ ++EK + ++E+SK S ++ NE RK M+Q++ +R R +S
Sbjct 22 VFTNAKAGMEGVDKEK--VQRVVYEMSKGSKYFQNEERKEALMKQKIEHMRDRCAKLSSL 79
Query 119 GGSAAASATARRVLQQLQQQQQRESGRVYVHLDMDMFFCAVE 160
S +R+ L+ + R+ R+++H+DMD F+ AVE
Sbjct 80 DLSNYQKVVDKRI---LELEATRDLSRIWLHVDMDAFYAAVE 118
> Hs7705344
Length=870
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 58/100 (58%), Gaps = 7/100 (7%)
Query 62 NSKAGMDMTEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQL-SVLRRRVQLFASSGG 120
++KAGM+ ++EK I I E +K S FY NE++K KQ+ Q++ ++++++ Q+ +S
Sbjct 23 DNKAGMEGLDKEK--INKIIMEATKGSRFYGNELKKEKQVNQRIENMMQQKAQI--TSQQ 78
Query 121 SAAASATARRVLQQLQQQQQRESGRVYVHLDMDMFFCAVE 160
A R +L +Q R VH+DMD F+ AVE
Sbjct 79 LRKAQLQVDRFAMEL--EQSRNLSNTIVHIDMDAFYAAVE 116
> CE00159
Length=518
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 23/103 (22%), Positives = 54/103 (52%), Gaps = 10/103 (9%)
Query 60 FANSKAGMDMTEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRRVQLFASSG 119
F ++KAGM+ ++EK K+ E + ++ + + ++ ++E+++ ++ R+Q
Sbjct 4 FNDNKAGMNGLDKEKI---TKVIEENTSASYSSFSKKQQSRIEEKVLEIKNRLQT----- 55
Query 120 GSAAASATARRVLQQLQQ--QQQRESGRVYVHLDMDMFFCAVE 160
+ + +++ L+ + R+ R V +DMD +F AVE
Sbjct 56 ATREERQKSEILMENLEMKLESSRDLSRDCVCIDMDAYFAAVE 98
> CE21230
Length=251
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 51/116 (43%), Gaps = 17/116 (14%)
Query 45 AAGPDLETVCRQTFIFANSKAGMDMTEEE------KAKIAAKIFELSKNSPFYANEMRKN 98
AAG DLE + + GM++ +EE K + + E+ KN
Sbjct 41 AAGNDLELNGARVLDLPTQEHGMELADEENPLGFVKDEAVGLVGEVEKNV---------- 90
Query 99 KQMEQQLSVLRRRVQLFASSGGSAAASATARRVLQQLQQQQQRESGRVYVHLDMDM 154
K+ +Q+S+ R +Q + GG AAA ++ V ++R + V +L+ D+
Sbjct 91 KKPAEQVSLNGRLLQP-VTDGGGAAAGSSDDVVYDYYAIHEKRGNPEVVGNLEQDI 145
> 7299037
Length=556
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 37/83 (44%), Gaps = 8/83 (9%)
Query 69 MTEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRRVQLFASSGGSAAASATA 128
+T EE + K LS N+PFY + ++ L LRR+ A S S + +
Sbjct 328 LTPEETQVLLQK---LSINNPFY-----RFRKQSNSLKELRRKTGSDAPSVRSRSETDLG 379
Query 129 RRVLQQLQQQQQRESGRVYVHLD 151
+ QQL QQ +V V LD
Sbjct 380 QVRGQQLMPHQQHSEAKVVVVLD 402
> SPAC2G11.12
Length=1328
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 5/57 (8%)
Query 34 SATAVPSSTSGAAGPDLETVCRQTFIFANSKAGMDMTEEEKAKIAAKIFELSKNSPF 90
SA+ PSS S D T Q + +D+ +EK++I+ +I E+ + PF
Sbjct 243 SASPTPSSVSSQISIDFSTWPHQNLL-----QYLDILRDEKSEISDRIIEVMERYPF 294
> Hs18598051
Length=1113
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 28/106 (26%), Positives = 43/106 (40%), Gaps = 11/106 (10%)
Query 14 RKNRAAAAGRATSANACEAVSATAVP---SSTSGAAGPDLETVCRQTFIFANSKAGMDM- 69
RK + AGR +S P ST PDLE V F+ SK +D+
Sbjct 618 RKLKLCKAGRPPKNTGKSLISTKNTPVSPGSTFPDVKPDLEDVDGVLFVSFESKEALDIH 677
Query 70 ----TEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRR 111
T EE + + A + +S + A + K++ + L LR +
Sbjct 678 AVDGTTEESSSLQAST---TNDSGYRARISQLEKELIEDLKTLRHK 720
> 7290581
Length=593
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 31 EAVSATAVPSSTSGAAGPDLETVCRQTFIFANSKAGMDMTEEEKAKIA 78
+++ A+ PS G A P++ R+ IF N KA T+++ +K+
Sbjct 535 KSLKASLKPSDRWGPANPEIR---REWVIFKNQKAAQRATQKDTSKLG 579
> SPCC553.07c
Length=493
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/91 (21%), Positives = 45/91 (49%), Gaps = 6/91 (6%)
Query 76 KIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRRVQLFASSGGSAAASATARRVLQQL 135
KI I+E SK S F+ E ++++++ ++ ++ V+ + S A + Q+
Sbjct 55 KINEIIYEASKGSKFFEAEQKRDRELRLRIEKVQVEVEKYQSKLRFDKAFQREWTIRQES 114
Query 136 QQ------QQQRESGRVYVHLDMDMFFCAVE 160
+ +R+ ++ VH+D D F+ ++E
Sbjct 115 VDTTVEDFRAKRDLTQIIVHVDCDAFYASIE 145
> CE21642
Length=374
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 32/83 (38%), Gaps = 2/83 (2%)
Query 70 TEEEKAKIAAKIFELSKNSPFYANEMRKNKQMEQQLSVLRRRVQLFASSGGSAAASATAR 129
EE ++ A + + K P++ K + L Q +G AA A
Sbjct 72 IHEESNEVGANVIAVQK-GPYFGTGNDKMMILSANYDTLEGN-QGVDDNGSGVAAVLEAA 129
Query 130 RVLQQLQQQQQRESGRVYVHLDM 152
RVL L R++ VYV DM
Sbjct 130 RVLSTLDNLYSRQNTIVYVFFDM 152
> SPBC23G7.12c
Length=403
Score = 27.7 bits (60), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 14/57 (24%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 26 SANACEAVSATAVPSSTSGAAGPDLETVCRQTFIFANSKAGMDMTEEEKAKIAAKIF 82
S N + ++ +GA+G +L+ VC + +FA + + +T+E+ AK+
Sbjct 332 SMNLTRGIDLKSIAEKMNGASGAELKGVCTEAGMFALRERRVHVTQEDFELAVAKVL 388
> 7300867
Length=229
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 12/65 (18%)
Query 50 LETVCRQTFIFANSKAGMDMTEEEKAKIAAK--IFELSKNSPFYANEMRKNKQMEQQLSV 107
LE +Q+ + + A E+E+ + AK +FE +K NKQ EQQ+SV
Sbjct 82 LEQSLKQSVMLGGTDAPKSRQEQEELLLKAKTLLFEKTKV----------NKQQEQQISV 131
Query 108 LRRRV 112
LR +V
Sbjct 132 LRTQV 136
Lambda K H
0.314 0.122 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40