bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1197_orf3
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
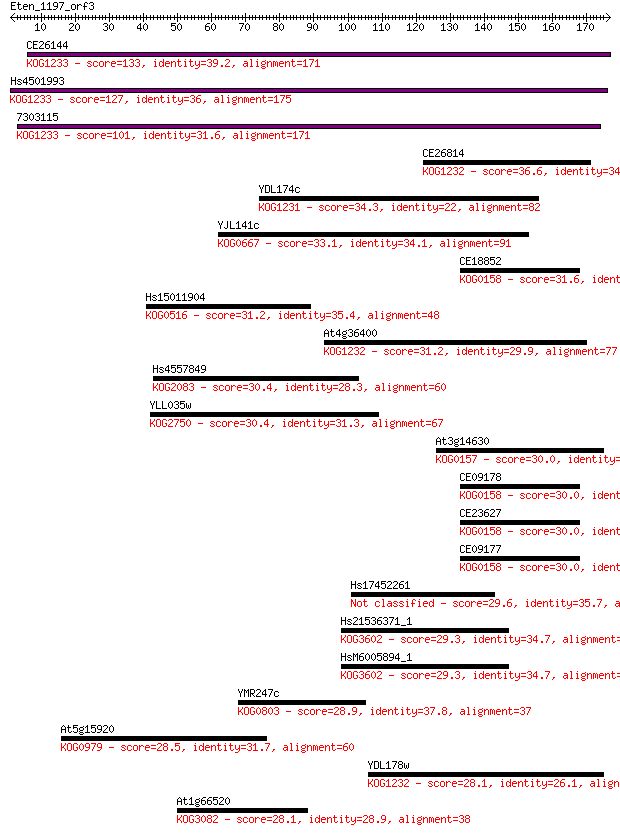
CE26144 133 2e-31
Hs4501993 127 1e-29
7303115 101 1e-21
CE26814 36.6 0.029
YDL174c 34.3 0.14
YJL141c 33.1 0.32
CE18852 31.6 0.96
Hs15011904 31.2 1.2
At4g36400 31.2 1.3
Hs4557849 30.4 1.8
YLL035w 30.4 2.1
At3g14630 30.0 2.3
CE09178 30.0 2.3
CE23627 30.0 2.7
CE09177 30.0 2.8
Hs17452261 29.6 3.3
Hs21536371_1 29.3 4.3
HsM6005894_1 29.3 4.4
YMR247c 28.9 6.0
At5g15920 28.5 7.2
YDL178w 28.1 9.6
At1g66520 28.1 9.8
> CE26144
Length=597
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 67/174 (38%), Positives = 101/174 (58%), Gaps = 7/174 (4%)
Query 6 KWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPAQ---D 62
KWNGWGY D+ +++ + G +Y + G M H+ + E+ GI+ F SPAQ D
Sbjct 22 KWNGWGYSDSQFAINKDGHVTFTGDKYEISGKVMPHFRPWFENYLGIDLGFVSPAQKLSD 81
Query 63 TLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDV 122
++ P V NE I+ LQ+ +I +N + RL GHGHT +++ LR G + RLPD+
Sbjct 82 VIIDAP--VENEDIIEFLQE--NKISFSNEARIRLMRGHGHTVHDMINLREGKIPRLPDI 137
Query 123 VLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLAF 176
V++PK ++ I+E A HN +IP GGGTSVT + P +E R V ++ +A
Sbjct 138 VVWPKSEHEIVKIIEGAMSHNCAIIPIGGGTSVTNALDTPETEKRAVISMDMAL 191
> Hs4501993
Length=658
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 63/177 (35%), Positives = 100/177 (56%), Gaps = 4/177 (2%)
Query 1 KVETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPA 60
+ E KWNGWGY D+ + I L G RY L G+ + + E++++ G+ + +
Sbjct 87 RQEVMKWNGWGYNDSKFIFNKKGQIELTGKRYPLSGMGLPTFKEWIQNTLGVNVEHKTTS 146
Query 61 QDTL--VIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVAR 118
+ +L P V NE F+ +L++ I + +R+F HGH +EIF LR G R
Sbjct 147 KASLNPSDTPPSVVNEDFLHDLKE--TNISYSQEADDRVFRAHGHCLHEIFLLREGMFER 204
Query 119 LPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSLA 175
+PD+VL+P H DV IV A ++N+C+IP GGGTSV+ G+ P E+R + ++ +
Sbjct 205 IPDIVLWPTCHDDVVKIVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTS 261
> 7303115
Length=631
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 54/171 (31%), Positives = 91/171 (53%), Gaps = 2/171 (1%)
Query 3 ETCKWNGWGYCDTYVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIEFAFSSPAQD 62
E KW GWGY D+ +I G +Y L G ++ + +++E + +
Sbjct 44 EALKWFGWGYNDSQF-YGKDGIICFRGEKYPLGGCELPSFTKWVEKKFDLRVDTTKQYPQ 102
Query 63 TLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDV 122
P+ V N F+ EL+ +++ + +RL HG T N+I+ L + R+PD+
Sbjct 103 LPRTYPRPVENAPFLHELKGTT-QVDYSAEGIDRLVRCHGQTLNDIYSLWHHKFRRIPDL 161
Query 123 VLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVS 173
V++P+ H +V +V A++HNV L+PFGGGTSV+ + P +ESRM+ +
Sbjct 162 VVWPRCHDEVVQLVRLANKHNVMLVPFGGGTSVSGAITCPQNESRMICALD 212
> CE26814
Length=487
Score = 36.6 bits (83), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 33/49 (67%), Gaps = 1/49 (2%)
Query 122 VVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVA 170
VVLYPK ++V AI+ S++ + ++P GG T + +G ++PV + +++
Sbjct 69 VVLYPKSTEEVSAILAYCSKNKLAVVPQGGNTGL-VGGSIPVHDEVVIS 116
> YDL174c
Length=587
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/82 (21%), Positives = 41/82 (50%), Gaps = 3/82 (3%)
Query 74 EAFIKELQQLAPEIEVANSEKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVE 133
+ +++L+Q+ S+ + H T F + + + P ++L+P ++V
Sbjct 108 DKVVEDLKQVLGNKPENYSDAKSDLDAHSDT---YFNTHHPSPEQRPRIILFPHTTEEVS 164
Query 134 AIVECASRHNVCLIPFGGGTSV 155
I++ +N+ ++PF GGTS+
Sbjct 165 KILKICHDNNMPVVPFSGGTSL 186
> YJL141c
Length=807
Score = 33.1 bits (74), Expect = 0.32, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 52/104 (50%), Gaps = 17/104 (16%)
Query 62 DTLVIPPQRV-----NNEAFIKELQQLAPEIEVANSEKERLFHGHGHTCNE--IFEL--- 111
DTL PP + N+ F+K +LAPE ++++K R+ C E I E
Sbjct 584 DTLGYPPSWMIDMGKNSGKFMK---KLAPEESSSSTQKHRM-KTIEEFCREYNIVEKPSK 639
Query 112 RYGTVARLPDVVL---YPKEHKDVEAIVECASRHNVCLIPFGGG 152
+Y +LPD++ YPK ++ + +++ ++ CLI F GG
Sbjct 640 QYFKWRKLPDIIRNYRYPKSIQNSQELIDQEMQNRECLIHFLGG 683
> CE18852
Length=534
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E + RHN+ IPFG G +G+ + +SE++
Sbjct 457 ERWLESSPRHNMSWIPFGAGPRQCVGMRLGLSEAK 491
> Hs15011904
Length=5430
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 41 HWHEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEIE 88
W E E + A SSP + T + Q +N+AF+ EL+Q +P+I+
Sbjct 2412 QWLESKEEVLKSMDAMSSPTK-TETVKAQAESNKAFLAELEQNSPKIQ 2458
> At4g36400
Length=524
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 9/77 (11%)
Query 93 EKERLFHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGG 152
+KERL T N + +Y ++L +L PK ++V I+E + ++P GG
Sbjct 83 DKERL-----ETANTDWMHKYKGSSKL---MLLPKNTQEVSQILEYCDSRRLAVVPQGGN 134
Query 153 TSVTLGVAVPVSESRMV 169
T + +G +VPV + +V
Sbjct 135 TGL-VGGSVPVFDEVIV 150
> Hs4557849
Length=1099
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 5/60 (8%)
Query 43 HEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEIEVANSEKERLFHGHG 102
H M+++P IE+ ++T I +VN + ++ +QLA + V S +R+ +G G
Sbjct 103 HNTMDAVPKIEYY-----RNTGSISGPKVNRPSLLEIHEQLAKNVAVTPSSADRVANGDG 157
> YLL035w
Length=632
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 6/69 (8%)
Query 42 WHEFMESIPGIEFAFSSPAQDTLVIPPQRVNNEAFIK--ELQQLAPEIEVANSEKERLFH 99
WH +SIP I+F+ + QDT +P NN I+ E + + V NS L
Sbjct 121 WHPLSQSIPTIDFSHFAGWQDTFFMPR---NNRFKIRDEEFKSFPCVLRVFNSNHTGLLE 177
Query 100 GHGHTCNEI 108
GH ++
Sbjct 178 A-GHLYRDV 185
> At3g14630
Length=508
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 126 PKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESRMVATVSL 174
P+ KD + A+++ VC +PFG G + +G + E++M + L
Sbjct 429 PERFKDG---IAKATKNQVCFLPFGWGPRICIGQNFALLEAKMALALIL 474
> CE09178
Length=515
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E +SRH + IPFG G +G+ + +SE++
Sbjct 421 ERWLEPSSRHTMSWIPFGAGPRQCVGMRLGLSEAK 455
> CE23627
Length=510
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E +SRH + IPFG G +G+ + +SE++
Sbjct 433 ERWLEPSSRHTMSWIPFGAGPRQCVGMRLGLSEAK 467
> CE09177
Length=527
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 133 EAIVECASRHNVCLIPFGGGTSVTLGVAVPVSESR 167
E +E +SRH + IPFG G +G+ + +SE++
Sbjct 433 ERWLEPSSRHTMSWIPFGAGPRQCVGMRLGLSEAK 467
> Hs17452261
Length=314
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query 101 HGHTCNEIFELRYGTVARLPDVVLYPKEHKDV-EAIVECASRH 142
+ +T +EI L YG V+ + + ++Y +K+V EA+ + SRH
Sbjct 266 NTNTSDEIIGLSYGVVSPMLNPIIYSLRNKEVKEAVKKVLSRH 308
> Hs21536371_1
Length=1436
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 98 FHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCL 146
H H T ++I L+ +A LPD+ K H V A + S N CL
Sbjct 34 LHQHVSTHSDILSLKNQCLATLPDLKTMEKPHGYVSAHPDILSLENQCL 82
> HsM6005894_1
Length=1436
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 98 FHGHGHTCNEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCL 146
H H T ++I L+ +A LPD+ K H V A + S N CL
Sbjct 34 LHQHVSTHSDILSLKNQCLATLPDLKTMEKPHGYVSAHPDILSLENQCL 82
> YMR247c
Length=1562
Score = 28.9 bits (63), Expect = 6.0, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 68 PQRVNNE--AFIKELQQLAPEIEVANSEKERLFHGHGHT 104
P+RVNN AFI + +L + + E L++ GHT
Sbjct 827 PERVNNHIVAFITVVSELVTDYNCLSEEPNDLYYDFGHT 865
> At5g15920
Length=1053
Score = 28.5 bits (62), Expect = 7.2, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 28/62 (45%), Gaps = 2/62 (3%)
Query 16 YVSMSSASMIRLHGSRYSLCGVDMGHWHEFMESIPGIE--FAFSSPAQDTLVIPPQRVNN 73
Y SSAS+ ++ SR LCGVD+G + +E F +L +R+
Sbjct 594 YGGHSSASVDSVYQSRLLLCGVDVGELEKLRSRKEELEDSILFMEETHKSLQTEQRRLEE 653
Query 74 EA 75
EA
Sbjct 654 EA 655
> YDL178w
Length=530
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 37/69 (53%), Gaps = 4/69 (5%)
Query 106 NEIFELRYGTVARLPDVVLYPKEHKDVEAIVECASRHNVCLIPFGGGTSVTLGVAVPVSE 165
NE + +Y ++L VL PK + V I+ + + ++P GG T + +G +VP+ +
Sbjct 91 NEDWMRKYKGQSKL---VLRPKSVEKVSLILNYCNDEKIAVVPQGGNTGL-VGGSVPIFD 146
Query 166 SRMVATVSL 174
+++ +L
Sbjct 147 ELILSLANL 155
> At1g66520
Length=355
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 50 PGIEFAFSSPAQDTLVIPPQRVNNEAFIKELQQLAPEI 87
P ++A L+ P++ +EAF+ L++L PE+
Sbjct 77 PVAQYALDKGLPSDLIFSPEKAGDEAFLSALRELQPEL 114
Lambda K H
0.320 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40