bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1157_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
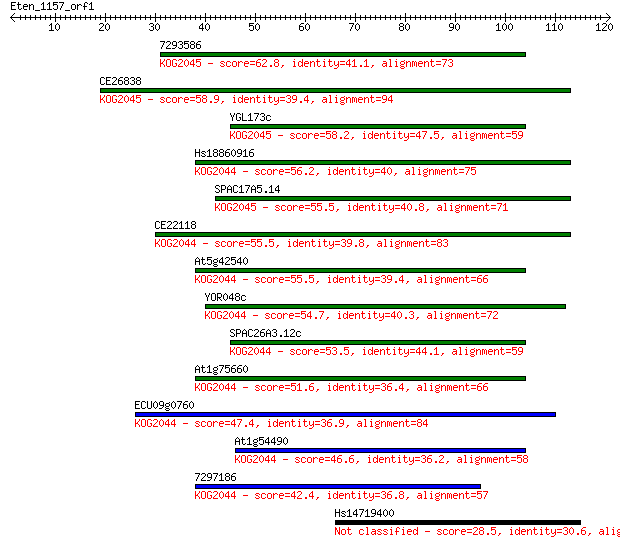
7293586 62.8 2e-10
CE26838 58.9 2e-09
YGL173c 58.2 4e-09
Hs18860916 56.2 1e-08
SPAC17A5.14 55.5 2e-08
CE22118 55.5 3e-08
At5g42540 55.5 3e-08
YOR048c 54.7 4e-08
SPAC26A3.12c 53.5 9e-08
At1g75660 51.6 4e-07
ECU09g0760 47.4 6e-06
At1g54490 46.6 1e-05
7297186 42.4 2e-04
Hs14719400 28.5 3.0
> 7293586
Length=1612
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 31 ASAATTAPSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGAD 90
A AA RE RFD+N I PGT FM + LR F+ K + +W ++ SG +
Sbjct 113 AKAAQRGELREHERFDSNCITPGTEFMVRLQEGLRAFLKTKISTDPLWQRC-TVILSGQE 171
Query 91 VPGEGEHKLMQLL 103
PGEGEHK+M +
Sbjct 172 APGEGEHKIMDYI 184
> CE26838
Length=1209
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 37/97 (38%), Positives = 47/97 (48%), Gaps = 6/97 (6%)
Query 19 AAAAAAGIAAAAASAATTAPSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVW 78
+A A A A PS + RFD+N I PGT+FM L +I K + W
Sbjct 118 SARTAHTQLAKALENGEIMPS--EARFDSNCITPGTYFMTRLHQKLDDWIKTKSATDSRW 175
Query 79 GAMRAIVFSGADVPGEGEHKLMQLLTR---GSWASPN 112
R I+ SG +VPGEGEHK+M + GS PN
Sbjct 176 QG-RKIILSGHNVPGEGEHKIMDFIRTERAGSSYDPN 211
> YGL173c
Length=1528
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 45 FDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKLMQLL 103
FD+NSI PGT FM L+ FI K + W ++ I+FSG +VPGEGEHK+M +
Sbjct 128 FDSNSITPGTEFMAKLTKNLQYFIHDKISNDSKWREVQ-IIFSGHEVPGEGEHKIMNFI 185
> Hs18860916
Length=950
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 39/75 (52%), Gaps = 2/75 (2%)
Query 38 PSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEH 97
P RFD+N I PGT FM LR +IA + + W + ++ S A PGEGEH
Sbjct 148 PEEIKERFDSNCITPGTEFMDNLAKCLRYYIADRLNNDPGWKNL-TVILSDASAPGEGEH 206
Query 98 KLMQLLTRGSWASPN 112
K+M + R A PN
Sbjct 207 KIMDYIRRQR-AQPN 220
> SPAC17A5.14
Length=1328
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 40/71 (56%), Gaps = 2/71 (2%)
Query 42 DNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKLMQ 101
+ +FD+N I PGT FM L FI K + W + ++FSG D PGEGEHK+M+
Sbjct 124 EEQFDSNCITPGTTFMERVSRQLYYFIHKKVTNDSQWQNIE-VIFSGHDCPGEGEHKIME 182
Query 102 LLTRGSWASPN 112
+ R A P+
Sbjct 183 YI-RTQKAQPS 192
> CE22118
Length=892
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/86 (38%), Positives = 45/86 (52%), Gaps = 4/86 (4%)
Query 30 AASAATTAPSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGA 89
A A +E+ FD+N I PGT FM LR +I + + W + I+ S A
Sbjct 141 AEGIAVPPKKKEEAHFDSNCITPGTPFMARLADALRYYIHDRVTNDASWANIE-IILSDA 199
Query 90 DVPGEGEHKLMQLL--TRGSWA-SPN 112
+VPGEGEHK+M + RG+ A PN
Sbjct 200 NVPGEGEHKIMDYVRKQRGNPAHDPN 225
> At5g42540
Length=1012
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 37/66 (56%), Gaps = 1/66 (1%)
Query 38 PSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEH 97
P + FD+N I PGT FM LR +I + S W ++ ++ S A+VPGEGEH
Sbjct 150 PKVDSQVFDSNVITPGTEFMATLSFALRYYIHVRLNSDPGWKNIK-VILSDANVPGEGEH 208
Query 98 KLMQLL 103
K+M +
Sbjct 209 KIMSYI 214
> YOR048c
Length=1006
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 2/72 (2%)
Query 40 REDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKL 99
R +D+N+I PGT FM + LR + A+K + W ++ I+ S A VPGEGEHK+
Sbjct 150 RNKKTWDSNAITPGTPFMDKLAAALRYWTAFKLATDPGWKNLQVII-SDATVPGEGEHKI 208
Query 100 MQLLTRGSWASP 111
M + R A P
Sbjct 209 MNFI-RSQRADP 219
> SPAC26A3.12c
Length=991
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 45 FDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKLMQLL 103
+D+N I PGT FM +LR +I K S W +R + S A VPGEGEHK+M+ +
Sbjct 157 WDSNCITPGTPFMDTLAKSLRYYIINKLNSDPCWRNVR-FILSDASVPGEGEHKIMEFI 214
> At1g75660
Length=985
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 1/66 (1%)
Query 38 PSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEH 97
P + FD+N I PGT FM V L+ ++ + W ++ ++ S A+VPGEGEH
Sbjct 148 PKVDSQVFDSNVITPGTEFMGVLSIALQYYVHLRLNHDVGWKNIK-VILSDANVPGEGEH 206
Query 98 KLMQLL 103
K+M +
Sbjct 207 KIMSYI 212
> ECU09g0760
Length=655
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 31/85 (36%), Positives = 44/85 (51%), Gaps = 2/85 (2%)
Query 26 IAAAAASAATTAPSREDNRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIV 85
IA + +++ A E +FD N I PGT FM L +I K S +R I+
Sbjct 146 IAEDPSESSSLAIDVETEKFDLNCITPGTPFMERLHKVLISYIQCKLSSDPEVSNLR-II 204
Query 86 FSGADVPGEGEHKLMQLL-TRGSWA 109
+S VPGEGE K+M + ++GS A
Sbjct 205 YSSYLVPGEGEQKIMSFIRSQGSIA 229
> At1g54490
Length=947
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 46 DTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKLMQLL 103
D+N I PGT FM + L+ +I + + W ++ ++ S ++VPGEGEHK+M +
Sbjct 157 DSNVITPGTPFMAILSVALQYYIQSRLNHNPGWRYVK-VILSDSNVPGEGEHKIMSYI 213
> 7297186
Length=885
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 38 PSRED-NRFDTNSIGPGTHFMFVAESTLRRFIAYKQRSSKVWGAMRAIVFSGADVPGE 94
P +E FD+N I PGT FM L F+ +Q ++ W ++ ++ S A+VPGE
Sbjct 149 PEKEKGEHFDSNCITPGTPFMDRLSKCLHYFVHDRQNNNPAWKGIK-VILSDANVPGE 205
> Hs14719400
Length=690
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 29/49 (59%), Gaps = 0/49 (0%)
Query 66 RFIAYKQRSSKVWGAMRAIVFSGADVPGEGEHKLMQLLTRGSWASPNAA 114
R +A+K +S + W A++A+ + G V + + K+++LL + A P A
Sbjct 48 RLLAHKIQSPQEWEALQALTYLGDRVSEKVKTKVIELLYSWTMALPEEA 96
Lambda K H
0.313 0.121 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40