bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1128_orf1
Length=274
Score E
Sequences producing significant alignments: (Bits) Value
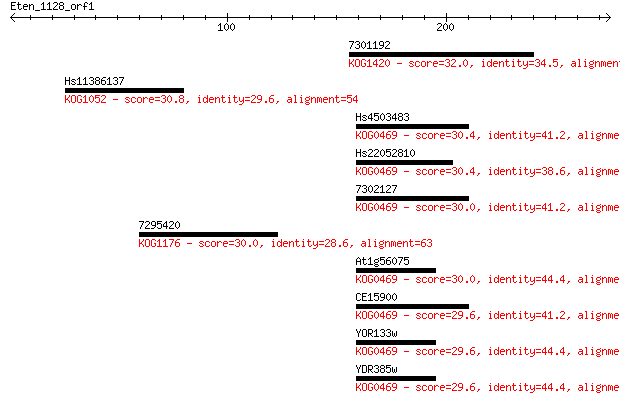
7301192 32.0 1.6
Hs11386137 30.8 3.0
Hs4503483 30.4 3.6
Hs22052810 30.4 3.7
7302127 30.0 4.8
7295420 30.0 4.8
At1g56075 30.0 5.2
CE15900 29.6 6.5
YOR133w 29.6 6.9
YDR385w 29.6 6.9
> 7301192
Length=1175
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 41/94 (43%), Gaps = 10/94 (10%)
Query 156 AFHLVEGLKVFAKSNPECAAVL----LQGGELDPITGEIHLNLYAEQVLESCSVTL---- 207
F L+ GL +FA S PE ++ GGEL G+ H+ + ES S L
Sbjct 319 VFFLLVGLAMFASSIPEIIELVGSGNKYGGELKREHGKRHIVVCGHITYESVSHFLKDFL 378
Query 208 -EERIAASL-ATGLRALPPQAEVAGRYQRRTATV 239
E+R + L PP E+ G ++R TV
Sbjct 379 HEDREDVDVEVVFLHRKPPDLELEGLFKRHFTTV 412
> Hs11386137
Length=908
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 24/54 (44%), Gaps = 0/54 (0%)
Query 26 VSFKKAKALEANFHLTSMSLKENGPLPATETLPAAAMQADQELKSRPTRIGRGL 79
++F K L +F L +SLKE G PA+ + + K +P I L
Sbjct 375 ITFNKTNGLRTDFDLDVISLKEEGLEKIGTWDPASGLNMTESQKGKPANITDSL 428
> Hs4503483
Length=858
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 9/52 (17%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNLYAEQVLESCSVTLEE 209
LVEGLK AKS+P ++ + GE + GE+H LE C LEE
Sbjct 520 LVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELH--------LEICLKDLEE 563
> Hs22052810
Length=846
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNLYAEQVLES 202
LVEGLK AKS+P ++ + GE + GE+HL + + + E
Sbjct 508 LVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELHLEICLKDLEED 552
> 7302127
Length=832
Score = 30.0 bits (66), Expect = 4.8, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 9/52 (17%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNLYAEQVLESCSVTLEE 209
LVEGLK AKS+P ++ + GE + GE+H LE C LEE
Sbjct 494 LVEGLKRLAKSDPMVQCIIEESGEHIIAGAGELH--------LEICLKDLEE 537
> 7295420
Length=564
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query 60 AAMQADQELKSRPTRIGRGLPLHERKVYQNLAR-NTPRVQGLTFDHNQIRWISYWKN-DQ 117
+A + +P +GR + + KV P V G HN +W Y+KN D+
Sbjct 365 SAGSVNLNFDEKPNSVGRAIRGIKIKVIDEQGEAQEPNVVGEICFHNSQKWAGYYKNPDE 424
Query 118 NKQVQ 122
+Q+Q
Sbjct 425 TRQIQ 429
> At1g56075
Length=855
Score = 30.0 bits (66), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNL 194
LVEGLK AKS+P + + GE + GE+HL +
Sbjct 516 LVEGLKRLAKSDPMVVCTMEESGEHIVAGAGELHLEI 552
> CE15900
Length=852
Score = 29.6 bits (65), Expect = 6.5, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 26/52 (50%), Gaps = 9/52 (17%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNLYAEQVLESCSVTLEE 209
LVEGLK AKS+P + + GE + GE+H LE C LEE
Sbjct 514 LVEGLKRLAKSDPMVQCIFEESGEHIIAGAGELH--------LEICLKDLEE 557
> YOR133w
Length=842
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNL 194
LVEGLK +KS+P + + GE + TGE+HL +
Sbjct 504 LVEGLKRLSKSDPCVLTYMSESGEHIVAGTGELHLEI 540
> YDR385w
Length=842
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 159 LVEGLKVFAKSNPECAAVLLQGGE-LDPITGEIHLNL 194
LVEGLK +KS+P + + GE + TGE+HL +
Sbjct 504 LVEGLKRLSKSDPCVLTYMSESGEHIVAGTGELHLEI 540
Lambda K H
0.318 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5914813682
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40