bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
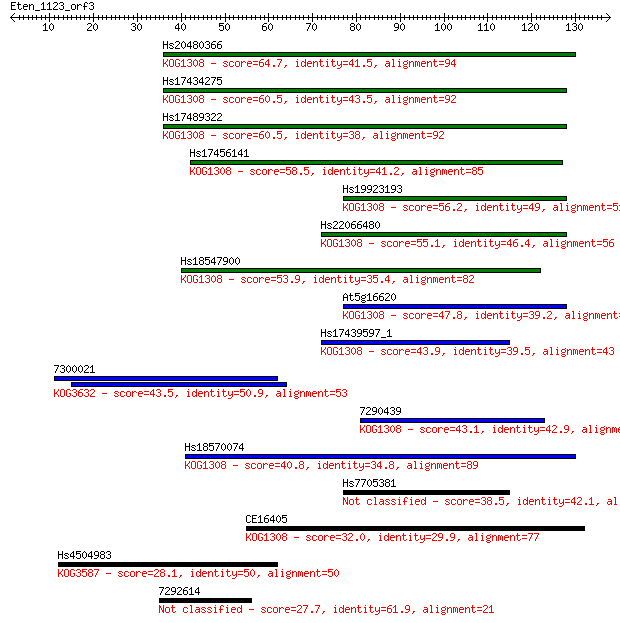
Query= Eten_1123_orf3
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
Hs20480366 64.7 4e-11
Hs17434275 60.5 9e-10
Hs17489322 60.5 1e-09
Hs17456141 58.5 4e-09
Hs19923193 56.2 2e-08
Hs22066480 55.1 3e-08
Hs18547900 53.9 8e-08
At5g16620 47.8 6e-06
Hs17439597_1 43.9 9e-05
7300021 43.5 1e-04
7290439 43.1 1e-04
Hs18570074 40.8 7e-04
Hs7705381 38.5 0.004
CE16405 32.0 0.37
Hs4504983 28.1 4.7
7292614 27.7 6.0
> Hs20480366
Length=369
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/95 (41%), Positives = 55/95 (57%), Gaps = 1/95 (1%)
Query 36 GGMSGSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSV-NDPDVQQVFSNPKMMAA 94
G GS GG PGG+P P GM G+ G L + +DP+V +P+++ A
Sbjct 275 GAQYGSFLGGFPGGMPANFPRGMPGMGGGMPGMAGMPGLNEILSDPEVLAAMQDPEVLVA 334
Query 95 FQDILTNPGNMAKYKDDPEVADAISKLMRKFGGGM 129
FQD+ NP NM+KY+ +P+V + ISKL KFGG +
Sbjct 335 FQDVAQNPANMSKYQSNPKVMNLISKLSVKFGGQV 369
> Hs17434275
Length=369
Score = 60.5 bits (145), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Query 36 GGMSGSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSV-NDPDVQQVFSNPKMMAA 94
G GS PGG PG +PG PGGM G+ G L + +DP+V +P++M A
Sbjct 275 GAQYGSFPGGFPGRMPGNFPGGMPGMGGGMPGMAGMPGLNEILSDPEVLAAMQDPEVMVA 334
Query 95 FQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
FQD+ NP +M+KY+ + +V + ISKL KFGG
Sbjct 335 FQDVAQNPASMSKYQSNSKVMNLISKLSAKFGG 367
> Hs17489322
Length=316
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 50/93 (53%), Gaps = 1/93 (1%)
Query 36 GGMSGSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAG-GLLGSVNDPDVQQVFSNPKMMAA 94
G S P G P GVPG P M G+ G GL ++DP++ +P++M A
Sbjct 222 GAQYCSFPSGFPAGVPGNCPRRMSGMGGGMAGMARIPGLNEILSDPEILAAMQDPEIMLA 281
Query 95 FQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
FQD+ +P NM+KY+ + + IS+L KFGG
Sbjct 282 FQDVAQDPANMSKYQRNTKTMHLISRLSAKFGG 314
> Hs17456141
Length=142
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/85 (41%), Positives = 51/85 (60%), Gaps = 7/85 (8%)
Query 42 MPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDILTN 101
MP GG+ G M G + G+ GM GL ++DP+V +PK++ AFQ ++ N
Sbjct 62 MPDNFHGGMLG-MGGAISGVAGMP------GLNEILSDPEVLAAKQDPKVIVAFQHVVQN 114
Query 102 PGNMAKYKDDPEVADAISKLMRKFG 126
P NM+KY+ +P+V + ISKL KFG
Sbjct 115 PANMSKYQSNPKVMNLISKLSAKFG 139
> Hs19923193
Length=369
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
++DP+V +P++M AFQD+ NP NM+KY+ +P+V + ISKL KFGG
Sbjct 317 LSDPEVLAAMQDPEVMVAFQDVAQNPANMSKYQSNPKVMNLISKLSAKFGG 367
> Hs22066480
Length=369
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 72 GLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
GL ++DP+ +P++M AFQD+ NP NM+KY+ +P+V + ISKL KFGG
Sbjct 312 GLNEILSDPEALAAMQDPEVMVAFQDVAQNPANMSKYQSNPKVMNLISKLSAKFGG 367
> Hs18547900
Length=105
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 43/82 (52%), Gaps = 14/82 (17%)
Query 40 GSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDIL 99
G MPG P G+ GM + G++ + +L + DP+V M AFQD+
Sbjct 27 GEMPGNFPRGI-----SGMARMPGLNEILSDLEVLADMQDPEV---------MVAFQDVA 72
Query 100 TNPGNMAKYKDDPEVADAISKL 121
NP N++KY +P+V + ISKL
Sbjct 73 QNPANLSKYHSNPKVMNLISKL 94
> At5g16620
Length=447
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
+ +PDV F NP++ AA + NP N+ KY++D EV D +K+ + F G
Sbjct 394 MENPDVAMAFQNPRVQAALMECSENPMNIMKYQNDKEVMDVFNKISQLFPG 444
> Hs17439597_1
Length=305
Score = 43.9 bits (102), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 72 GLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
GL ++DP+V +P++M AFQD+ NP N++KY+ +P++
Sbjct 254 GLNEILSDPEVLTAMQDPEVMVAFQDVAQNPANISKYQSNPKI 296
> 7300021
Length=1251
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/51 (50%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 11 PGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMGGL 61
PG PG MPG PG+M+G G PG M G MPG + G +PG M G G L
Sbjct 932 PGQMPGQMPGQMPGQMAGQMAGQMPGQMPGQMPGQMSGQMPGQMMGPRGPL 982
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 15 PGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMGGLEG 63
PG MPG PG+M G G G M G MPG +PG + G MPG M G G
Sbjct 932 PGQMPGQMPGQMPGQMAGQMAGQMPGQMPGQMPGQMSGQMPGQMMGPRG 980
> 7290439
Length=373
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 81 DVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 122
D+ S+P++ AA QDIL+NPGN+ KY +P++ + I K++
Sbjct 295 DILGAMSDPEVSAAIQDILSNPGNITKYASNPKIYNLIKKIV 336
> Hs18570074
Length=369
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 46/89 (51%), Gaps = 11/89 (12%)
Query 41 SMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDILT 100
+ P G+PG G F L +DP+V +P++M AFQD+
Sbjct 292 NFPRGMPGMGGGMSGMAGMP-------GFNEIL----SDPEVLAAMQDPEVMVAFQDVAQ 340
Query 101 NPGNMAKYKDDPEVADAISKLMRKFGGGM 129
NP N++KY+ +P+V + ISKL KFGG +
Sbjct 341 NPANVSKYQSNPKVMNLISKLSAKFGGQV 369
> Hs7705381
Length=404
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
+++P VQ +NPK + AF+D+L NP N ++ +DPE
Sbjct 350 LDNPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPET 387
> CE16405
Length=422
Score = 32.0 bits (71), Expect = 0.37, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 33/77 (42%), Gaps = 26/77 (33%)
Query 55 PGGMGGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
P ++ +P+++ A DI+ NP NM KY ++P+V
Sbjct 327 PEIAAAIQ-------------------------DPEVLPALMDIMQNPANMMKYINNPKV 361
Query 115 ADAISKLMRKFGGGMAG 131
A ISKL K G GM G
Sbjct 362 AKLISKLQSK-GAGMPG 377
> Hs4504983
Length=250
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 28/56 (50%), Gaps = 6/56 (10%)
Query 12 GGFPGG-MPGGFPGEMS-GGYPGGAPGGMSGSMPGGIPG----GVPGGMPGGMGGL 61
GG+PG PG +PG+ G YPG AP G PG PG GV G P G G
Sbjct 34 GGYPGASYPGAYPGQAPPGAYPGQAPPGAYHGAPGAYPGAPAPGVYPGPPSGPGAY 89
> 7292614
Length=167
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 35 PGGMSGSMPGGIPGGVPGGMP 55
PG MSG+MPG +PG + GMP
Sbjct 90 PGAMSGAMPGAMPGAMSCGMP 110
Lambda K H
0.316 0.144 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40