bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1113_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
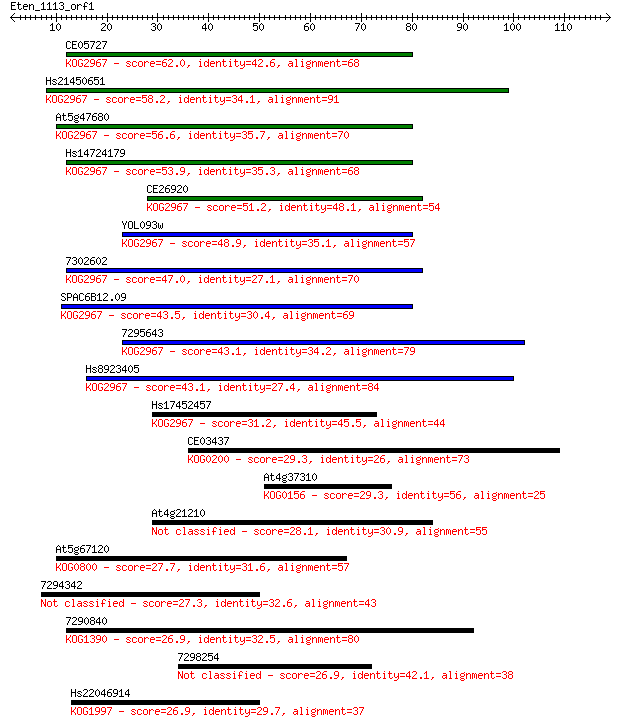
CE05727 62.0 3e-10
Hs21450651 58.2 4e-09
At5g47680 56.6 1e-08
Hs14724179 53.9 7e-08
CE26920 51.2 4e-07
YOL093w 48.9 2e-06
7302602 47.0 8e-06
SPAC6B12.09 43.5 1e-04
7295643 43.1 1e-04
Hs8923405 43.1 1e-04
Hs17452457 31.2 0.42
CE03437 29.3 1.9
At4g37310 29.3 1.9
At4g21210 28.1 4.0
At5g67120 27.7 5.3
7294342 27.3 6.5
7290840 26.9 8.2
7298254 26.9 8.9
Hs22046914 26.9 10.0
> CE05727
Length=297
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 41/68 (60%), Gaps = 0/68 (0%)
Query 12 VHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELG 71
+H H+E F P+EIV L+ ++ L D+D +KV+VIGG+VD KGL + A+E G
Sbjct 151 IHLHQEKLENLFKPEEIVYLTSESENVLSDLDDTKVYVIGGIVDHNSQKGLCYRIAQEKG 210
Query 72 LTCRKLPF 79
KLP
Sbjct 211 FGHAKLPL 218
> Hs21450651
Length=319
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 55/92 (59%), Gaps = 3/92 (3%)
Query 8 NSWLVHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQA 67
+S+L+ EE + F + +V L+PD+ L DVD +KV+++GGLVD +I K ++F +A
Sbjct 188 SSYLLDITEEDCFSLFPLETLVYLTPDSEHALEDVDLNKVYILGGLVDESIQKKVTFQKA 247
Query 68 EELGLTCRKLPFKK-LGRSSRGCASKSHARVL 98
E + +LP ++ + R+ G H+ +L
Sbjct 248 REYSVKTARLPIQEYMVRNQNG--KNYHSEIL 277
> At5g47680
Length=344
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 42/72 (58%), Gaps = 2/72 (2%)
Query 10 WLVHRHEEPYWQCFS--PDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQA 67
W + + Y + + D +V L+ D+ L D+DP +++IGGLVDR KG++ ++A
Sbjct 180 WFIEKESRCYIEAMADQKDNLVYLTADSETVLDDLDPKHIYIIGGLVDRNRFKGITMTKA 239
Query 68 EELGLTCRKLPF 79
+E G+ KLP
Sbjct 240 QEQGIKTAKLPI 251
> Hs14724179
Length=339
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 0/68 (0%)
Query 12 VHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELG 71
+H E Y + ++++ L+ D+ L ++D SK +VIGGLVD KGL++ QA + G
Sbjct 167 IHIKPEHYSELIKKEDLIYLTSDSPNILKELDESKAYVIGGLVDHNHHKGLTYKQASDYG 226
Query 72 LTCRKLPF 79
+ +LP
Sbjct 227 INHAQLPL 234
> CE26920
Length=370
Score = 51.2 bits (121), Expect = 4e-07, Method: Composition-based stats.
Identities = 26/58 (44%), Positives = 36/58 (62%), Gaps = 4/58 (6%)
Query 28 IVVLSPDASEELLDVDPSKVFVIGGLVDRT----ISKGLSFSQAEELGLTCRKLPFKK 81
++ LSPDA+EEL VD K++VIGG+VDR I K S ++ G+ RKLP +
Sbjct 234 VIYLSPDATEELESVDDDKIYVIGGIVDRVPEPGIPKHASLEASQSAGVFARKLPIDR 291
> YOL093w
Length=293
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 23 FSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGLTCRKLPF 79
S D+IV L+ D E+L ++P +++GG+VD+ K L +A+++G+ R+LP
Sbjct 178 ISKDKIVYLTADTEEKLEKLEPGMRYIVGGIVDKNRYKELCLKKAQKMGIPTRRLPI 234
> 7302602
Length=446
Score = 47.0 bits (110), Expect = 8e-06, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 12 VHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELG 71
++ H + + + F +V L+P E+L+ +P ++++G +VD ++ LS ++A+ LG
Sbjct 260 INLHSKCFTELFPKQNLVYLTPHCREDLVTYNPDDIYIVGAMVDTMNNEPLSLAKAKRLG 319
Query 72 LTCRKLPFKK 81
L +LP +
Sbjct 320 LRMARLPLDR 329
> SPAC6B12.09
Length=304
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 42/82 (51%), Gaps = 13/82 (15%)
Query 11 LVHRHEEPYWQCFSP-------------DEIVVLSPDASEELLDVDPSKVFVIGGLVDRT 57
V + ++ W+ ++P +++V LS D+ + ++D K+++IG +VD+
Sbjct 150 FVLKGQQNNWKRYNPTTKSYLEEFESQKEKLVYLSADSDNTITELDEDKIYIIGAIVDKN 209
Query 58 ISKGLSFSQAEELGLTCRKLPF 79
K L ++A E G+ KLP
Sbjct 210 RYKNLCQNKASEQGIKTAKLPI 231
> 7295643
Length=319
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 40/80 (50%), Gaps = 1/80 (1%)
Query 23 FSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGLTCRKLPF-KK 81
F ++V L+ ++ L + P +VIGGLVD KGL S+A GLT +LP +
Sbjct 173 FEHSQLVYLTCESDRVLDKLQPGCTYVIGGLVDHNHFKGLCHSRATSAGLTTARLPLSEH 232
Query 82 LGRSSRGCASKSHARVLRSK 101
+ +R S H L +K
Sbjct 233 VDMKTRAVLSTYHVFELLTK 252
> Hs8923405
Length=252
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 42/84 (50%), Gaps = 0/84 (0%)
Query 16 EEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGLTCR 75
E+ + F D I+ L+ D+ + KV+VIG VD+++ G S ++A+ L L
Sbjct 124 EKSHVDLFPKDSIIYLTADSPNVMTTFRHDKVYVIGSFVDKSMQPGTSLAKAKRLNLATE 183
Query 76 KLPFKKLGRSSRGCASKSHARVLR 99
LP K + G + + +++R
Sbjct 184 CLPLDKYLQWEIGNKNLTLDQMIR 207
> Hs17452457
Length=127
Score = 31.2 bits (69), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 10/54 (18%)
Query 29 VVLSP---DASEE-------LLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGL 72
V L+P D SE+ L DVD ++V + GLVD +I K ++F +A+E
Sbjct 55 VYLTPRPLDYSEQTCGTEYTLEDVDVNEVCIFDGLVDESIQKKVTFQKAQEYSF 108
> CE03437
Length=1227
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 19/82 (23%), Positives = 40/82 (48%), Gaps = 9/82 (10%)
Query 36 SEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGLTC-----RKLPFKKLGRSSRGCA 90
SE+ +++P+K+ +GG DR + ++F++ +L + R + KL + G
Sbjct 503 SEQFQEIEPNKLVSVGGTFDRGLIYNVTFTEGMDLKCSSIRKGNRAITVNKLIKIGEGFE 562
Query 91 SKSH----ARVLRSKTTKNIKE 108
+ H ++ S+ +K I E
Sbjct 563 VRPHHEKSSKATESEPSKIIYE 584
> At4g37310
Length=518
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 51 GGLVDRTISKGLSFSQAEELGLTCR 75
GG VD T GLS +AE L +TCR
Sbjct 474 GGQVDMTEGPGLSLPKAEPLVVTCR 498
> At4g21210
Length=403
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 33/56 (58%), Gaps = 3/56 (5%)
Query 29 VVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELGL-TCRKLPFKKLG 83
+V D + L ++DP KVF G +++ + +G+ ++A+ LGL + K + +LG
Sbjct 294 IVNGVDLPKTLFEIDPRKVF--GLMINPLVLQGIREARAKSLGLGSSFKTKYSELG 347
> At5g67120
Length=272
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Query 10 WLVHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQ 66
WL+ + W +SP + +SPD + LDV+ + G + RTIS +F Q
Sbjct 19 WLLFGDDWDLWDTYSPVDADDISPDPT---LDVNGDGPAIEPGSLLRTISWETTFEQ 72
> 7294342
Length=285
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 20/43 (46%), Gaps = 0/43 (0%)
Query 7 GNSWLVHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFV 49
N L HR E QC D ++V + + S EL +D +V
Sbjct 205 ANEHLEHRLIEEIRQCAKEDAVIVYNENGSGELHTIDQEDYYV 247
> 7290840
Length=410
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 37/80 (46%), Gaps = 10/80 (12%)
Query 12 VHRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELG 71
V + + W+ +VVL A+ + LDVDP+KV V GG V +I + S A +
Sbjct 325 VRKEDVAMWEVNEAFSLVVL---ANIKKLDVDPAKVNVHGGAV--SIGHPIGMSGARLVA 379
Query 72 LTCRKLPFKKLGRSSRGCAS 91
L +L GCAS
Sbjct 380 HLSHSLKKGEL-----GCAS 394
> 7298254
Length=1379
Score = 26.9 bits (58), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 34 DASEELLDVDPSKVFVIGGLVDRTISKGLSFSQAEELG 71
D +EE+ D P KV V GL D+ IS + F +A+ L
Sbjct 421 DGTEEMNDEFPEKVEVSKGLEDKNIS-SVVFDEADALN 457
> Hs22046914
Length=1573
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 11/37 (29%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 13 HRHEEPYWQCFSPDEIVVLSPDASEELLDVDPSKVFV 49
HR E Y QCF + + V+ + +DP+K ++
Sbjct 1301 HRLEAFYGQCFGAEFVEVIKDSTPVDKTKLDPNKAYI 1337
Lambda K H
0.315 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40