bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1106_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
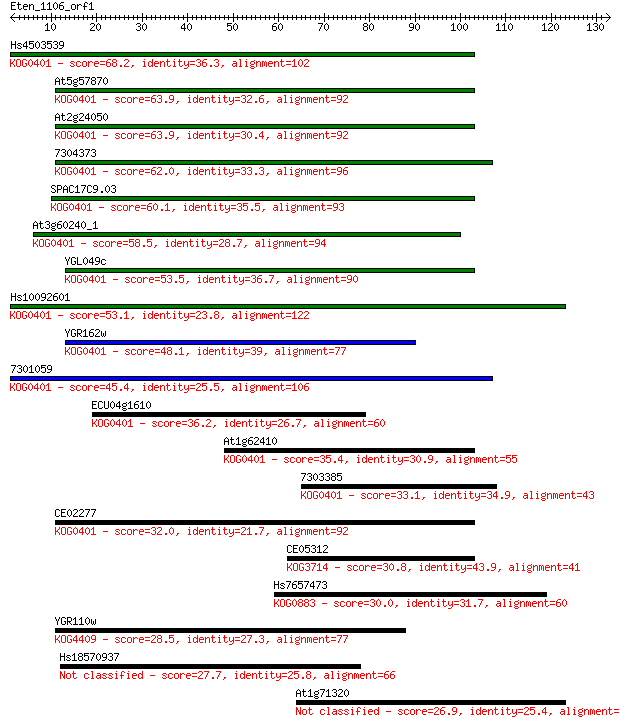
Hs4503539 68.2 4e-12
At5g57870 63.9 7e-11
At2g24050 63.9 8e-11
7304373 62.0 3e-10
SPAC17C9.03 60.1 1e-09
At3g60240_1 58.5 3e-09
YGL049c 53.5 1e-07
Hs10092601 53.1 1e-07
YGR162w 48.1 4e-06
7301059 45.4 3e-05
ECU04g1610 36.2 0.017
At1g62410 35.4 0.031
7303385 33.1 0.13
CE02277 32.0 0.32
CE05312 30.8 0.74
Hs7657473 30.0 1.2
YGR110w 28.5 3.3
Hs18570937 27.7 5.5
At1g71320 26.9 9.4
> Hs4503539
Length=907
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 64/109 (58%), Gaps = 10/109 (9%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
EK++ DA + R ++G+LNKLT E+FD++ ++LN G++ + +K ++ +I DKA+ +
Sbjct 70 EKERHDA---IFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDKALEEP 126
Query 61 HFIQMYVMLCGKLKTDFQ-------ELLQDDHRSTKFRRILIDQCEDSF 102
+ +Y LC +L D E +ST FRR+LI + +D F
Sbjct 127 KYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEF 175
> At5g57870
Length=780
Score = 63.9 bits (154), Expect = 7e-11, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 55/94 (58%), Gaps = 2/94 (2%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
+L+ +KG+LNKLT E++D + Q++ +GI +K ++ +IFDKAV + F MY LC
Sbjct 215 VLKTVKGILNKLTPEKYDLLKGQLIESGITSADILKGVITLIFDKAVLEPTFCPMYAKLC 274
Query 71 GKLKTDFQEL--LQDDHRSTKFRRILIDQCEDSF 102
+ + + F+R+L++ C+++F
Sbjct 275 SDINDQLPTFPPAEPGDKEITFKRVLLNICQEAF 308
> At2g24050
Length=747
Score = 63.9 bits (154), Expect = 8e-11, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 59/94 (62%), Gaps = 2/94 (2%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
+++ +KG+LNKLT E+++ + Q+++AGI +K ++Q+IF+ A+ Q F +MY +LC
Sbjct 174 VVKSVKGILNKLTPEKYELLKGQLIDAGITSADILKEVIQLIFENAILQPTFCEMYALLC 233
Query 71 GKLKTDFQELLQDD--HRSTKFRRILIDQCEDSF 102
+ ++ + F+R+L++ C+++F
Sbjct 234 FDINGQLPSFPSEEPGGKEITFKRVLLNNCQEAF 267
> 7304373
Length=1666
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 59/101 (58%), Gaps = 5/101 (4%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLC 70
L+R ++G+LNKLT ERFD + ++I+ I +V ++ ++F+KA+ + +F Y LC
Sbjct 749 LVRRVRGILNKLTPERFDTLVEEIIKLKIDTPDKVDEVIVLVFEKAIDEPNFSVSYARLC 808
Query 71 GKLKTDFQ---ELLQDDHRSTK--FRRILIDQCEDSFNANL 106
+L + + E ++ + +S FR L+D+ E F N+
Sbjct 809 QRLAAEVKVIDERMESETKSNSAHFRNALLDKTEQEFTQNV 849
> SPAC17C9.03
Length=1403
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 33/108 (30%), Positives = 58/108 (53%), Gaps = 15/108 (13%)
Query 10 VLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKE-----VKALMQMIFDKAVSQHHFIQ 64
V+ R +KG LNK+T+E+FD+I QIL ++ KE +K ++Q+ F+KA + +F
Sbjct 1007 VVQRKVKGSLNKMTLEKFDKISDQILEIAMQSRKENDGRTLKQVIQLTFEKATDEPNFSN 1066
Query 65 MYVMLCGKLKTDFQELLQDDHRSTK----------FRRILIDQCEDSF 102
MY K+ + ++D+ K FR+ L+ +C++ F
Sbjct 1067 MYARFARKMMDSIDDSIRDEGVLDKNNQPVRGGLLFRKYLLSRCQEDF 1114
> At3g60240_1
Length=1528
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 53/94 (56%), Gaps = 1/94 (1%)
Query 6 DAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQM 65
D + R LK +LNKLT + F+++++Q+ + I + ++ IFDKA+ + F +M
Sbjct 907 DEEQAKQRQLKSILNKLTPQNFEKLFEQVKSVNIDNAVTLSGVISQIFDKALMEPTFCEM 966
Query 66 YVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCE 99
Y C L + ++ + T F+R+L+++C+
Sbjct 967 YADFCFHLSGALPDFNENGEKIT-FKRLLLNKCQ 999
> YGL049c
Length=914
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/100 (33%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Query 13 RHLKGLLNKLTIERFDRIYQQILNAGIKQEKE-----VKALMQMIFDKAVSQHHFIQMYV 67
R +K LLNKLT+E FD I +IL+ + + E +K +++ IF KA + H+ MY
Sbjct 568 RKMKSLLNKLTLEMFDSISSEILDIANQSKWEDDGETLKIVIEQIFHKACDEPHWSSMYA 627
Query 68 MLCGKLKTDFQELLQDDHRSTK-----FRRILIDQCEDSF 102
LCGK+ D ++D K L+ +C + F
Sbjct 628 QLCGKVVKDLDPNIKDKENEGKNGPKLVLHYLVARCHEEF 667
> Hs10092601
Length=1585
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/126 (23%), Positives = 65/126 (51%), Gaps = 13/126 (10%)
Query 1 EKQKEDAQMV----LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKA 56
+ Q +D + + L R ++ +LNKLT + F+++ +Q+ + E+ +K ++ ++F+KA
Sbjct 740 DSQADDPENIKTQELFRKVRSILNKLTPQMFNQLMKQVSGLTVDTEERLKGVIDLVFEKA 799
Query 57 VSQHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLS 116
+ + F Y +C L T + + FR++L+++C+ F + +
Sbjct 800 IDEPSFSVAYANMCRCLVTLKVPMADKPGNTVNFRKLLLNRCQKEFEKD---------KA 850
Query 117 EDDAYE 122
+DD +E
Sbjct 851 DDDVFE 856
> YGR162w
Length=952
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 46/82 (56%), Gaps = 5/82 (6%)
Query 13 RHLKGLLNKLTIERFDRIYQQIL---NAGIKQE--KEVKALMQMIFDKAVSQHHFIQMYV 67
R +K LLNKLT+E FD I +IL N + + + +KA+++ IF KA + H+ MY
Sbjct 608 RKMKSLLNKLTLEMFDAISSEILAIANISVWETNGETLKAVIEQIFLKACDEPHWSSMYA 667
Query 68 MLCGKLKTDFQELLQDDHRSTK 89
LCGK+ + + D+ K
Sbjct 668 QLCGKVVKELNPDITDETNEGK 689
> 7301059
Length=1905
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/106 (25%), Positives = 59/106 (55%), Gaps = 6/106 (5%)
Query 1 EKQKEDAQMVLLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQH 60
+ + +D + VL + ++G+LNKLT + F+ + +++ + + E ++ +M +IF+K +S+
Sbjct 1097 QDEDDDVEGVL-KKVRGILNKLTPDNFEVLLKEMSSIKMDNEAKMTNVMLLIFEKTISEP 1155
Query 61 HFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANL 106
+F Y C K F E+ ++ + F LI + + F +N+
Sbjct 1156 NFAPTYARFC---KVLFHEIKAEN--KSLFTSSLITRIQHEFESNV 1196
> ECU04g1610
Length=627
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 19 LNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQ 78
N+LT + + + + + +E+K + +++FDKA+S+ F++ Y +L LK ++Q
Sbjct 210 FNRLTAKNIGLVIKNLKAIRVGTIEEMKEIAKILFDKAISEPTFVKYYALLVLDLKKEWQ 269
> At1g62410
Length=223
Score = 35.4 bits (80), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 48 LMQMIFDKAVSQHHFIQMYVMLCGKLKTDFQELLQDDHRS--TKFRRILIDQCEDSF 102
L +IFDKAV + F MY LC ++ ++ F+R+L++ C+ F
Sbjct 13 LTTLIFDKAVLEPTFCPMYAQLCFDIRHKMPRFPPSAPKTDEISFKRVLLNTCQKVF 69
> 7303385
Length=869
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 65 MYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLE 107
MY LC +L + ++ S+ F R+LI C D FN L+
Sbjct 1 MYAQLCKRLSEEAPSFDKEPSNSSTFLRLLIAVCRDKFNNRLK 43
> CE02277
Length=1156
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/93 (21%), Positives = 51/93 (54%), Gaps = 4/93 (4%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQ-EKEVKALMQMIFDKAVSQHHFIQMYVML 69
+ + ++ L+NK+T + ++ ++ + + ++ +++++FDKAV + F +Y +
Sbjct 517 VCKKVRSLMNKVTPTSQRPLTEEFISYNVSSNDAQLAQVVEIVFDKAVEEPKFCALYAEM 576
Query 70 CGKLKTDFQELLQDDHRSTKFRRILIDQCEDSF 102
C K + + EL Q +S FR ++ + + +F
Sbjct 577 C-KAQAN-HELSQTGGKSA-FRNKVLTRTQMTF 606
> CE05312
Length=651
Score = 30.8 bits (68), Expect = 0.74, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 62 FIQMYVMLCGKLKTDFQEL-LQDDHRSTKFRRILIDQCEDSF 102
FI+ + LC DF EL + DD R+T FR D+ E SF
Sbjct 418 FIEDFSFLCTSTCVDFVELKISDDLRNTGFRFCCYDKPEISF 459
> Hs7657473
Length=520
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 27/60 (45%), Gaps = 2/60 (3%)
Query 59 QHHFIQMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSED 118
QH +MY+ C + T F + D T FRR+ D C S + P+ P G+ D
Sbjct 5 QHQKDKMYIT-CAEY-THFYGGKKPDLPQTNFRRLPFDHCSLSLQPFVYPVCTPDGIVFD 62
> YGR110w
Length=445
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 40/80 (50%), Gaps = 3/80 (3%)
Query 11 LLRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQ---MIFDKAVSQHHFIQMYV 67
L+R+ + L +K+T R ++ ILN +Q K +AL + IF+K S + + +
Sbjct 295 LVRNFRQLGSKITSGWSYRRFKHILNGDPEQSKRFEALHRYAYAIFNKRGSGEYLLSFAL 354
Query 68 MLCGKLKTDFQELLQDDHRS 87
G+ + ++ L D +S
Sbjct 355 KCGGEPRLSLEQQLFDGKKS 374
> Hs18570937
Length=214
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 32/66 (48%), Gaps = 0/66 (0%)
Query 12 LRHLKGLLNKLTIERFDRIYQQILNAGIKQEKEVKALMQMIFDKAVSQHHFIQMYVMLCG 71
L L+G +NKL IE R+++++LN + +E L + D ++ + + L G
Sbjct 141 LMRLRGDINKLKIEHTCRLHRRMLNDATYELEERDELADLFCDSPLASSFSLSTPLKLIG 200
Query 72 KLKTDF 77
K +
Sbjct 201 VTKMNI 206
> At1g71320
Length=392
Score = 26.9 bits (58), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 15/59 (25%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 64 QMYVMLCGKLKTDFQELLQDDHRSTKFRRILIDQCEDSFNANLEPIAVPQGLSEDDAYE 122
++ + KL +LL D + +FR IL+ +C+ + N+E + + L D E
Sbjct 254 SLFWLTLKKLSQTSYQLLAIDLHTEEFRWILLPECDTKYATNIEMWNLNERLCLSDVLE 312
Lambda K H
0.322 0.137 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40