bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1105_orf1
Length=338
Score E
Sequences producing significant alignments: (Bits) Value
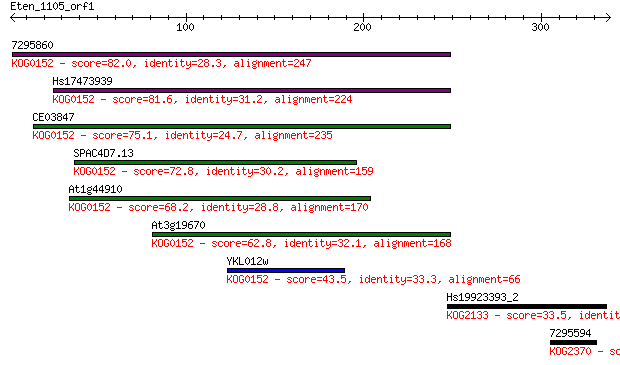
7295860 82.0 2e-15
Hs17473939 81.6 2e-15
CE03847 75.1 2e-13
SPAC4D7.13 72.8 9e-13
At1g44910 68.2 2e-11
At3g19670 62.8 1e-09
YKL012w 43.5 7e-04
Hs19923393_2 33.5 0.71
7295594 31.2 3.0
> 7295860
Length=806
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 70/256 (27%), Positives = 129/256 (50%), Gaps = 11/256 (4%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
L++ +++ + Y + F WT + E +R + +++ +F+ + K++ + ++ +
Sbjct 307 LMSSDKMNSQMKYFRCEEVFAGTRTWTAVPEPDRRDIYEDCIFNLAKREKEEARLLKKRN 366
Query 62 VAMLEEILEQNPTEFAFQKRWSEVREQLFSHPKLQN------MMKIDVLQVWEEWVR--- 112
+ +L E+LE T WSE + L + +N M K D L V+EE +R
Sbjct 367 MKVLGELLESM-TSINHATTWSEAQVMLLDNVAFKNDVTLLGMDKEDALIVFEEHIRTLE 425
Query 113 HGYEQERKQRRAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAM 172
++ER++ + ++ R++RK RD+F LL ++G+L+S S WV + D R+ AM
Sbjct 426 KEEDEEREREKKRMKRQQRKNRDSFLALLDSLHEEGKLTSMSLWVELYPIISADLRFSAM 485
Query 173 VGQSGSTPRELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDE 232
+GQSGSTP +LF V+ L+ + K + + ++ K SF F V
Sbjct 486 LGQSGSTPLDLFKFYVENLKARFHDEKKIIREILKEKAFVVQAKT-SFEDFATVVCEDKR 544
Query 233 VKNLNMLHVKLVFESL 248
+L+ +VKL + SL
Sbjct 545 SASLDAGNVKLTYNSL 560
> Hs17473939
Length=569
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 70/233 (30%), Positives = 112/233 (48%), Gaps = 11/233 (4%)
Query 25 EWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSE 84
E W + E +R E + + +F + K++ KQ RR ++ L+ IL+ + FQ WS+
Sbjct 76 EVWAVVPERDRKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKSILD-GMSSVNFQTTWSQ 134
Query 85 VREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQRRAQIFRRER---KRRD 135
++ L +P +LQNM K D L +EE +R +E ++R R R K R+
Sbjct 135 AQQYLMDNPSFAQDHQLQNMDKEDALICFEEHIRALEREEEEERERARLRERRQQRKNRE 194
Query 136 AFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELFSDAVQQLQQQH 195
AF+ L E + G+L S S W+ V D R+ M+GQ GSTP +LF V++L+ +
Sbjct 195 AFQTFLDELHETGQLHSMSTWMELYPAVSTDVRFANMLGQPGSTPLDLFKFYVEELKARF 254
Query 196 ERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLVFESL 248
K K G + + +F F + L+ ++KL F SL
Sbjct 255 HDEKKIIKDILKDRGFCV-EVNTAFEDFAHVISFDKRAAALDAGNIKLTFNSL 306
> CE03847
Length=724
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 58/244 (23%), Positives = 123/244 (50%), Gaps = 11/244 (4%)
Query 14 YIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNP 73
Y +D F E W + + +R E F++ + + K+K ++ R+ D+A +L Q+
Sbjct 318 YQKASDIFSKEPLWIAVNDEDRKEIFRDCIDFVARRDKEKKEEDRKRDIAAFSHVL-QSM 376
Query 74 TEFAFQKRWSEVREQLFSHPK------LQNMMKIDVLQVWEEWVRHGYEQERKQRRA--- 124
+ ++ W++ + L+ +P+ L M K D L V+E+ ++ ++ +++
Sbjct 377 EQITYKTTWAQAQRILYENPQFAERKDLHFMDKEDALTVFEDHIKQAEKEHDEEKEQEEK 436
Query 125 QIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELF 184
++ R++RK R+ +R LL+ +GEL+S S W + D+R+ M+ Q GS+P +LF
Sbjct 437 RLRRQQRKVREEYRLLLESLHKRGELTSMSLWTSLFPIISTDTRFELMLFQPGSSPLDLF 496
Query 185 SDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLV 244
V+ L++Q+ + K +G + + +F + V ++ ++ ++KL
Sbjct 497 KFFVEDLKEQYTEDRRLIKEILTEKGCQVI-ATTEYREFSDWVVSHEKGGKVDHGNMKLC 555
Query 245 FESL 248
+ SL
Sbjct 556 YNSL 559
> SPAC4D7.13
Length=695
Score = 72.8 bits (177), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 48/168 (28%), Positives = 92/168 (54%), Gaps = 15/168 (8%)
Query 37 EFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSEVREQLFSHPK-- 94
E+ Q+ + D +Q+ KD+ K+ + ++L + F RWS + + P+
Sbjct 309 EYKQKLLEDEKQLEKDRRKEALDDFCSLLRNM------NFEPYTRWSVAQAKFDQDPRYT 362
Query 95 ----LQNMMKIDVLQVWEEWVRH---GYEQERKQRRAQIFRRERKRRDAFRELLQEAVDK 147
++ + K+D L +E+ V+H Y ++++++ + R ERK RDAFR LLQ+ +
Sbjct 363 RNSNMKYLSKLDALVAFEDHVKHLEREYILDKQKQKKEKHRIERKNRDAFRALLQDLRVQ 422
Query 148 GELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELFSDAVQQLQQQH 195
+++ +++W ++ D RYL ++GQSGSTP +LF D + L+ +
Sbjct 423 KKITLRTKWKELYPIIKDDPRYLNLLGQSGSTPLDLFWDTIVDLENMY 470
> At1g44910
Length=946
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/174 (28%), Positives = 96/174 (55%), Gaps = 5/174 (2%)
Query 34 ERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSEVREQLFSHP 93
+R++ F ++ + E+ ++K + R+ +A + LE +W +++++L
Sbjct 512 DREDLFDNYIVELERKEREKAAEEHRQYMADYRKFLETC-DYIKAGTQWRKIQDRLEDDD 570
Query 94 KLQNMMKIDVLQVWEEWVRHGYEQERKQRRA---QIFRRERKRRDAFRELLQEAVDKGEL 150
+ + KID L +EE++ ++E + +R + R ERK RDAFR LL+E V G L
Sbjct 571 RCSCLEKIDRLIGFEEYILDLEKEEEELKRVEKEHVRRAERKNRDAFRTLLEEHVAAGIL 630
Query 151 SSKSEWVGFARRVEKDSRYLAMVGQ-SGSTPRELFSDAVQQLQQQHERLKHFFK 203
++K+ W+ + ++ +Y A+ SGSTP++LF D ++L++Q+ K + K
Sbjct 631 TAKTYWLDYCIELKDLPQYQAVASNTSGSTPKDLFEDVTEELEKQYHEDKSYVK 684
> At3g19670
Length=960
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 54/179 (30%), Positives = 98/179 (54%), Gaps = 19/179 (10%)
Query 81 RWSEVREQLFSHPKLQNMMKIDVLQVWEEWVR---HGYEQERKQRRAQIFRRERKRRDAF 137
+W +V+++L + + KID L++++E++R E+++K ++ ++ + ERK RD F
Sbjct 575 QWRKVQDRLEVDERCSRLEKIDQLEIFQEYLRDLEREEEEKKKIQKEELKKVERKHRDEF 634
Query 138 RELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQ-SGSTPRELFSDAVQQLQQQHE 196
LL E + GEL++K+ W + +V+ Y A+ SG+TP++LF DAV+ L+++
Sbjct 635 HGLLDEHIATGELTAKTIWRDYLMKVKDLPVYSAIASNSSGATPKDLFEDAVEDLKKRDH 694
Query 197 RLKHFFKRFADREGLDLRDKALS----FTQFYETVR---GLDEVKNLNMLHVKLVFESL 248
LK K L LR LS F +F ++ G + ++ + KLVF+ L
Sbjct 695 ELKSQIKDV-----LKLRKVNLSAGSTFDEFKVSISEDIGFPLIPDVRL---KLVFDDL 745
> YKL012w
Length=583
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Query 123 RAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRE 182
R + + R+R RD F+ LL+E K + + + W ++ D R+L M+G++GS+ +
Sbjct 348 RLRNYTRDRIARDNFKSLLREVPIK--IKANTRWSDIYPHIKSDPRFLHMLGRNGSSCLD 405
Query 183 LFSDAV 188
LF D V
Sbjct 406 LFLDFV 411
> Hs19923393_2
Length=1458
Score = 33.5 bits (75), Expect = 0.71, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 43/100 (43%), Gaps = 10/100 (10%)
Query 247 SLKNADTKPAAAAAAAADGKQRSSRSSRTDPDAGAYRSSKHDDATSSSSSSSSSSA---- 302
S++ ++ + + + KQ +S RT P RSS + +++S+SS+ S A
Sbjct 463 SMRTRRSRGSMSTLRSGRKKQPASPDGRTSPINEDIRSSGRNSPSAASTSSNDSKAETVK 522
Query 303 ------NHHGSHRDREREREREREKEREREKEREREKKKK 336
S + +R+RE+ E +R KK K
Sbjct 523 KSAKKVKEEASSPLKSNKRQREKVASDTEEADRTSSKKTK 562
> 7295594
Length=720
Score = 31.2 bits (69), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 22/26 (84%), Gaps = 0/26 (0%)
Query 305 HGSHRDREREREREREKEREREKERE 330
H HRD+ER++ REREK++ R+K+RE
Sbjct 45 HRDHRDKERDQRREREKDKSRDKKRE 70
Lambda K H
0.315 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8059015174
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40