bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1088_orf1
Length=146
Score E
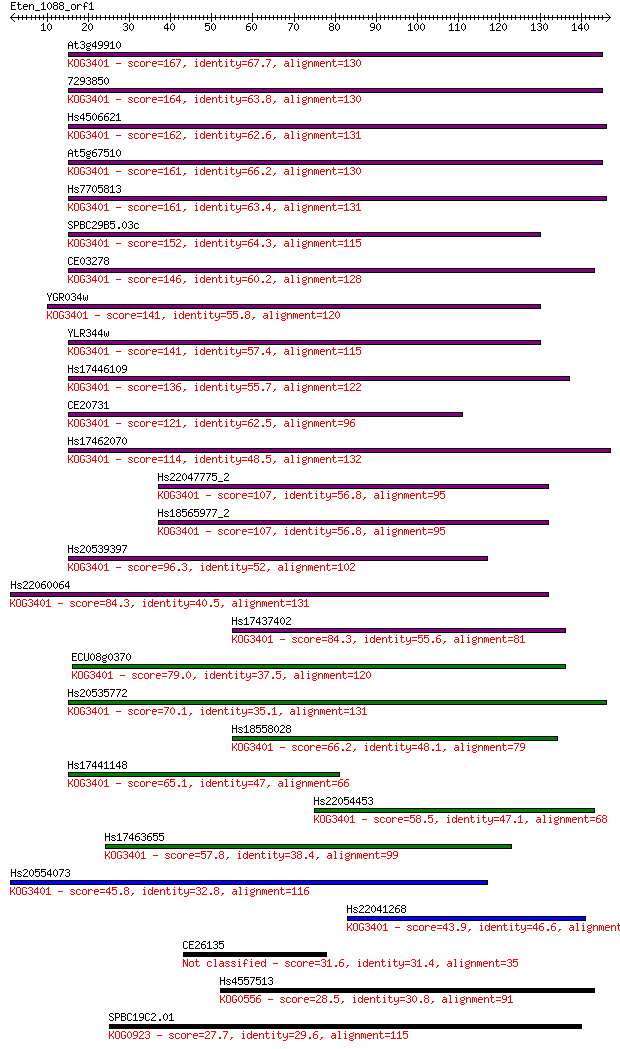
Sequences producing significant alignments: (Bits) Value
At3g49910 167 5e-42
7293850 164 4e-41
Hs4506621 162 3e-40
At5g67510 161 3e-40
Hs7705813 161 5e-40
SPBC29B5.03c 152 2e-37
CE03278 146 2e-35
YGR034w 141 4e-34
YLR344w 141 5e-34
Hs17446109 136 2e-32
CE20731 121 6e-28
Hs17462070 114 9e-26
Hs22047775_2 107 1e-23
Hs18565977_2 107 1e-23
Hs20539397 96.3 2e-20
Hs22060064 84.3 7e-17
Hs17437402 84.3 8e-17
ECU08g0370 79.0 3e-15
Hs20535772 70.1 1e-12
Hs18558028 66.2 2e-11
Hs17441148 65.1 5e-11
Hs22054453 58.5 4e-09
Hs17463655 57.8 7e-09
Hs20554073 45.8 3e-05
Hs22041268 43.9 1e-04
CE26135 31.6 0.53
Hs4557513 28.5 4.2
SPBC19C2.01 27.7 7.2
> At3g49910
Length=146
Score = 167 bits (424), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 88/137 (64%), Positives = 109/137 (79%), Gaps = 7/137 (5%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++SRRK RKAHF A SS RR IM++PL +LR+K+NVRS+PIRKDDEV IVRG + RE
Sbjct 7 VTSSRRKNRKAHFTASSSERRVIMSSPLSTDLRQKYNVRSMPIRKDDEVQIVRGTYKGRE 66
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGK--- 130
GKV QVYR+K+VIHIERI+REK NG ++ +GI PSKVVITK +L+KDRK+LL RK K
Sbjct 67 GKVVQVYRRKWVIHIERITREKVNGTTVNVGIQPSKVVITKLRLDKDRKSLLERKAKGRA 126
Query 131 AANK---GKYTEKDVAQ 144
AA+K K+T +DV Q
Sbjct 127 AADKEKGTKFTSEDVMQ 143
> 7293850
Length=149
Score = 164 bits (416), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 83/138 (60%), Positives = 108/138 (78%), Gaps = 8/138 (5%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
VS+SRRK RK HF APS +RR++M+APL KELR+K+NVRS+PIR+DDEV ++RG F +
Sbjct 7 VSSSRRKNRKRHFQAPSHIRRRLMSAPLSKELRQKYNVRSMPIRRDDEVQVIRGHFKGNQ 66
Query 75 -GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGK-- 130
GKV Q YRKKFV+++E+I RE ANG ++ +GIHPSKV+I K KL+KDRKA+L R+GK
Sbjct 67 VGKVVQAYRKKFVVYVEKIQRENANGTNVYVGIHPSKVLIVKLKLDKDRKAILERRGKGR 126
Query 131 ----AANKGKYTEKDVAQ 144
+KGKYTE+ AQ
Sbjct 127 LAALGKDKGKYTEETAAQ 144
> Hs4506621
Length=145
Score = 162 bits (409), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 82/137 (59%), Positives = 108/137 (78%), Gaps = 6/137 (4%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++ R K RK HF APS +RRKIM++PL KELR+K+NVRS+PIRKDDEV +VRG + ++
Sbjct 7 VTSDRSKNRKRHFNAPSHIRRKIMSSPLSKELRQKYNVRSMPIRKDDEVQVVRGHYKGQQ 66
Query 75 -GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA- 131
GKV QVYRKK+VI+IER+ REKANG ++ +GIHPSKVVIT+ KL+KDRK +L RK K+
Sbjct 67 IGKVVQVYRKKYVIYIERVQREKANGTTVHVGIHPSKVVITRLKLDKDRKKILERKAKSR 126
Query 132 ---ANKGKYTEKDVAQV 145
KGKY E+ + ++
Sbjct 127 QVGKEKGKYKEETIEKM 143
> At5g67510
Length=146
Score = 161 bits (408), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 86/137 (62%), Positives = 110/137 (80%), Gaps = 7/137 (5%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
VS+SRRK RKAHF APSSVRR +M++PL K+LR K NVRS+PIRKDDEV +VRG F RE
Sbjct 7 VSSSRRKNRKAHFTAPSSVRRVLMSSPLSKDLRNKHNVRSMPIRKDDEVQVVRGTFKGRE 66
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARK--GKA 131
GKV QVYR+K+VIHIERI+REK NG ++ +G++ S V+ITK +L+KDRK+LL RK G+A
Sbjct 67 GKVMQVYRRKWVIHIERITREKVNGSTVNVGVNASNVMITKLRLDKDRKSLLERKANGRA 126
Query 132 A---NKG-KYTEKDVAQ 144
A KG K++ +DV +
Sbjct 127 AADKEKGTKFSAEDVME 143
> Hs7705813
Length=145
Score = 161 bits (407), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 83/137 (60%), Positives = 108/137 (78%), Gaps = 6/137 (4%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++ R K RK HF APS VRRKIM++PL KELR+K+NVRS+PIRKDDEV +VRG + ++
Sbjct 7 VTSDRSKNRKRHFNAPSHVRRKIMSSPLSKELRQKYNVRSMPIRKDDEVQVVRGHYKGQQ 66
Query 75 -GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA- 131
GKV QVYRKK+VI+IER+ REKANG ++ +GIHPSKVVIT+ KL+KDRK +L RK K+
Sbjct 67 IGKVVQVYRKKYVIYIERVQREKANGTTVHVGIHPSKVVITRLKLDKDRKKILERKAKSR 126
Query 132 ---ANKGKYTEKDVAQV 145
KGKY E+ + ++
Sbjct 127 QVGKEKGKYKEELIEKM 143
> SPBC29B5.03c
Length=126
Score = 152 bits (384), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 74/116 (63%), Positives = 96/116 (82%), Gaps = 1/116 (0%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++SRRK RKAHFGAPSSVRR +M+APL KELR+++ +RSLP+R+DD++ ++RG RE
Sbjct 7 VTSSRRKQRKAHFGAPSSVRRVLMSAPLSKELREQYKIRSLPVRRDDQITVIRGSNKGRE 66
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKG 129
GK+T VYRKKF++ IER++REKANG S +GI SKVVITK L+KDRK L+ RKG
Sbjct 67 GKITSVYRKKFLLLIERVTREKANGASAPVGIDASKVVITKLHLDKDRKDLIVRKG 122
> CE03278
Length=142
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 77/135 (57%), Positives = 101/135 (74%), Gaps = 7/135 (5%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
VS+ K+RKAHF APS RR+IM+APL KELR K +R++PIR DDEV+++RGR
Sbjct 7 VSSDSGKSRKAHFNAPSHERRRIMSAPLTKELRTKHGIRAIPIRTDDEVVVMRGRHKGNT 66
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKAAN 133
G+V + YRKKFVIHI++I+REKANG ++ IGIHPSKV ITK KL+KDR+AL+ RK +
Sbjct 67 GRVLRCYRKKFVIHIDKITREKANGSTVHIGIHPSKVAITKLKLDKDRRALVERKAAGRS 126
Query 134 ------KGKYTEKDV 142
KGK+T++ V
Sbjct 127 RVTGILKGKHTDETV 141
> YGR034w
Length=129
Score = 141 bits (356), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 67/121 (55%), Positives = 95/121 (78%), Gaps = 1/121 (0%)
Query 10 CMHSGVSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGR 69
C VS+ RRKARKA+F APSS RR +++APL KELR ++ +++LPIR+DDEV++VRG
Sbjct 5 CKTMYVSSDRRKARKAYFTAPSSERRVLLSAPLSKELRAQYGIKALPIRRDDEVLVVRGS 64
Query 70 FHDREGKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARK 128
+EGK++ VYR KF + ++++++EK NG S+ I +HPSK+VITK L+KDRKAL+ RK
Sbjct 65 KKGQEGKISSVYRLKFAVQVDKVTKEKVNGASVPINLHPSKLVITKLHLDKDRKALIQRK 124
Query 129 G 129
G
Sbjct 125 G 125
> YLR344w
Length=127
Score = 141 bits (355), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 66/116 (56%), Positives = 94/116 (81%), Gaps = 1/116 (0%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
VS+ RRKARKA+F APSS RR +++APL KELR ++ +++LPIR+DDEV++VRG +E
Sbjct 8 VSSDRRKARKAYFTAPSSQRRVLLSAPLSKELRAQYGIKALPIRRDDEVLVVRGSKKGQE 67
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKG 129
GK++ VYR KF + ++++++EK NG S+ I +HPSK+VITK L+KDRKAL+ RKG
Sbjct 68 GKISSVYRLKFAVQVDKVTKEKVNGASVPINLHPSKLVITKLHLDKDRKALIQRKG 123
> Hs17446109
Length=135
Score = 136 bits (342), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 68/124 (54%), Positives = 93/124 (75%), Gaps = 2/124 (1%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++++ K K HF APS + RKIM++P+ KELR+K+NV+S+PIRKDDEV +VRG + ++
Sbjct 7 VTSNQIKNSKRHFNAPSHIHRKIMSSPVSKELRQKYNVQSMPIRKDDEVQVVRGHYKGQQ 66
Query 75 -GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKAA 132
GKV QVYRKK+VI+IE + REK NG ++ GIHP KV IT+ KL+KD K +L K K+
Sbjct 67 IGKVVQVYRKKYVIYIEWVQREKTNGTTVHTGIHPRKVAITRLKLDKDSKKILEWKAKSR 126
Query 133 NKGK 136
GK
Sbjct 127 QVGK 130
> CE20731
Length=106
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 60/97 (61%), Positives = 77/97 (79%), Gaps = 1/97 (1%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
VS+ K+RKAHF APS RR+IM+APL KELR K +R++PIR DDEV+++RGR
Sbjct 7 VSSDSGKSRKAHFNAPSHERRRIMSAPLTKELRTKHGIRAIPIRTDDEVVVMRGRHKGNT 66
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKV 110
G+V + YRKKFVIHI++I+REKANG ++ IGIHPSKV
Sbjct 67 GRVLRCYRKKFVIHIDKITREKANGSTVHIGIHPSKV 103
> Hs17462070
Length=140
Score = 114 bits (284), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 64/138 (46%), Positives = 92/138 (66%), Gaps = 11/138 (7%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++ R K K HF APS RRKIM++PL KELR+K+NVRS I+KDDEV + RG ++
Sbjct 7 VTSDRSKNCKRHFNAPSHTRRKIMSSPLSKELRQKYNVRSKTIQKDDEVQVARGHCKGQQ 66
Query 75 -GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA- 131
G++ QVYRKK+ I+IER+ EKAN ++ +GIHP+++ KL+K K +L RK K+
Sbjct 67 IGQIIQVYRKKYAIYIERVQWEKANSIAVPVGIHPARL-----KLDKGHKKILERKAKSR 121
Query 132 ---ANKGKYTEKDVAQVD 146
GKY ++ + +V
Sbjct 122 QVGKENGKYKKETIGKVQ 139
> Hs22047775_2
Length=98
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/97 (55%), Positives = 75/97 (77%), Gaps = 2/97 (2%)
Query 37 IMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE-GKVTQVYRKKFVIHIERISRE 95
I ++ LCKELR+K+NV+S+ IRKDDEV +V + ++ GKV QVYRKK+V +I ++ +E
Sbjct 2 IRSSSLCKELRQKYNVQSMTIRKDDEVQVVHRHYKGQQIGKVVQVYRKKYVTYIAQVQQE 61
Query 96 KANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA 131
KANG ++ IGIHPSKVV T+ KL+KD K +L RK K+
Sbjct 62 KANGTTVHIGIHPSKVVSTRLKLDKDCKKILERKAKS 98
> Hs18565977_2
Length=98
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/97 (55%), Positives = 75/97 (77%), Gaps = 2/97 (2%)
Query 37 IMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE-GKVTQVYRKKFVIHIERISRE 95
I ++ LCKELR+K+NV+S+ IRKDDEV +V + ++ GKV QVYRKK+V +I ++ +E
Sbjct 2 IRSSSLCKELRQKYNVQSMTIRKDDEVQVVHRHYKGQQIGKVVQVYRKKYVTYIAQVQQE 61
Query 96 KANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA 131
KANG ++ IGIHPSKVV T+ KL+KD K +L RK K+
Sbjct 62 KANGTTVHIGIHPSKVVSTRLKLDKDCKKILERKAKS 98
> Hs20539397
Length=137
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 53/106 (50%), Positives = 71/106 (66%), Gaps = 8/106 (7%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++ R K RK HF APS +RRKIM++PL +ELR+K+NVRS+ E M+ G + D
Sbjct 7 VTSDRSKNRKRHFKAPSHIRRKIMSSPLSEELRQKYNVRSMA----SERMMKLGLYQDTT 62
Query 75 ---GKVTQVYRKKFVIHIERISREKANGESLI-GIHPSKVVITKPK 116
GK VYRKK+V +IE + REKANG ++ GIHPSK++ K K
Sbjct 63 KQIGKAVLVYRKKYVTYIEWVQREKANGSAVHGGIHPSKILEWKAK 108
> Hs22060064
Length=124
Score = 84.3 bits (207), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 53/131 (40%), Positives = 69/131 (52%), Gaps = 32/131 (24%)
Query 1 HFSQSAAALCMHSGVSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKD 60
H AA + + V++ + K HF APS +RRKI+++ L KELR K+NVRS+PIRKD
Sbjct 21 HSEAGAAQMKFNPFVTSDQSNNHKRHFNAPSHIRRKILSSLLSKELRDKYNVRSMPIRKD 80
Query 61 DEVMIVRGRFHDREGKVTQVYRKKFVIHIERISREKANGESLIGIHPSKVVITKPKLNKD 120
D Q + +H GIHPSKVVIT+ KL+KD
Sbjct 81 D----------------VQAFTP--TVH--------------TGIHPSKVVITRLKLDKD 108
Query 121 RKALLARKGKA 131
RK LL RK K+
Sbjct 109 RKMLLERKAKS 119
> Hs17437402
Length=101
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 45/85 (52%), Positives = 61/85 (71%), Gaps = 4/85 (4%)
Query 55 LPIRKDDEVMIVRGRFH---DREGKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKV 110
+P RKD+EV +VR R H + G V QVYRKK+VI+IE + +EKANG ++ +GIHPSK
Sbjct 1 MPNRKDNEVQVVRVRGHYKGQQIGTVVQVYRKKYVIYIEWVQQEKANGTTVHVGIHPSKA 60
Query 111 VITKPKLNKDRKALLARKGKAANKG 135
VIT+ KL+KDRK + +K K+ G
Sbjct 61 VITRLKLDKDRKKIPEQKAKSHQAG 85
> ECU08g0370
Length=143
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/121 (37%), Positives = 72/121 (59%), Gaps = 1/121 (0%)
Query 16 SASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDREG 75
++SRRK RKAHF +R +A L +ELRK++ R+ P+R D V++ G+F +EG
Sbjct 9 TSSRRKQRKAHFACNDGEKRIRSSARLSRELRKEYGFRTFPLRTGDTVVVKGGKFDGKEG 68
Query 76 KVTQVYRKKFVIHIERISREKANG-ESLIGIHPSKVVITKPKLNKDRKALLARKGKAANK 134
++++ + + ++IE K +G +L+ I+PSK+ ITK L RK L RK +A
Sbjct 69 VISKINYRDYKVYIEGCFVTKNDGTNALVPIYPSKLTITKFFLENGRKEQLERKKRAREH 128
Query 135 G 135
G
Sbjct 129 G 129
> Hs20535772
Length=104
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 70/136 (51%), Gaps = 45/136 (33%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V++ R K K+HF APS +RRKI+++ L KELR+K+ V+
Sbjct 7 VTSDRSKNHKSHFSAPSHIRRKILSSSLSKELRQKYEVQW-------------------- 46
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA-- 131
EKANG ++ +GIHP+KVVIT+ KL+KD K +L ++ K+
Sbjct 47 --------------------EKANGTTVHVGIHPNKVVITRLKLDKDCKKILEKEAKSRQ 86
Query 132 --ANKGKYTEKDVAQV 145
KGKY E+ + ++
Sbjct 87 VGKEKGKYKEETIEKM 102
> Hs18558028
Length=98
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/81 (46%), Positives = 56/81 (69%), Gaps = 3/81 (3%)
Query 55 LPIRKDDEVMIVRGRFHDRE-GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVI 112
+ IRKDD V V+G ++ G V +VYRKK VI+ E + ++KANG ++ +GIHPSKVVI
Sbjct 1 MTIRKDD-VQTVQGHCKGQQIGHVLKVYRKKDVIYTEWVQQQKANGTTVRVGIHPSKVVI 59
Query 113 TKPKLNKDRKALLARKGKAAN 133
T+ K++KD K + +K K+
Sbjct 60 TRLKVDKDHKKIHEQKAKSCQ 80
> Hs17441148
Length=126
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 49/67 (73%), Gaps = 1/67 (1%)
Query 15 VSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE 74
V + K K+HF APS + +KIM++PL KELR+K NV+S+ I+KD++V +V+G + ++
Sbjct 60 VICEQTKNWKSHFNAPSHIHKKIMSSPLSKELRQKRNVQSVFIQKDNKVQVVQGHYKGQQ 119
Query 75 -GKVTQV 80
GK+ QV
Sbjct 120 IGKIVQV 126
> Hs22054453
Length=462
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 49/73 (67%), Gaps = 5/73 (6%)
Query 75 GKVTQVYRKKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRKALLARKGKA-- 131
GKV Q+YR+K+VI+IE + REKA+ ++ + PSK +I++ KL K+ K +LA K K+
Sbjct 385 GKVVQIYREKYVIYIEWVQREKADDTTVRVDFRPSKGIISRLKLGKEHKKILAWKAKSHQ 444
Query 132 --ANKGKYTEKDV 142
KGKY E+ +
Sbjct 445 VGKEKGKYKEETI 457
> Hs17463655
Length=132
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 53/100 (53%), Gaps = 27/100 (27%)
Query 24 KAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLPIRKDDEVMIVRGRFHDREGKVTQVYRK 83
K HF APS + RKIM++ L KEL K+N RF K
Sbjct 56 KKHFNAPSHICRKIMSSSLSKELDLKYN----------------SRFTG----------K 89
Query 84 KFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRK 122
K+VI+IE + +EKAN ++ +GI+ SKVV+T L++D K
Sbjct 90 KYVIYIEWVQQEKANSTTVHVGIYSSKVVVTGLTLDEDHK 129
> Hs20554073
Length=184
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 57/122 (46%), Gaps = 11/122 (9%)
Query 1 HFSQSA----AALCMHSGVSASRRKARKAHFGAPSSVRRKIMTAPLCKELRKKFNVRSLP 56
H+S A + H V++ RK K HF S RKI++ KEL + N S+P
Sbjct 44 HYSNVADTVLVTINFHPLVTSDSRKNSKHHFSVLSPRHRKILSFQFFKELWQGSNDHSIP 103
Query 57 IRKDDEVMIVRGRFHD-REGKVTQVY-RKKFVIHIERISREKANGESLIGIHPSKVVITK 114
I K F D + G QV+ ++ IHI+R+ E N ++ IH S +I++
Sbjct 104 IHKRSSRW-----FEDTKRGDNGQVWLTERICIHIKRVQHENVNRVTVHVIHHSTGLISE 158
Query 115 PK 116
K
Sbjct 159 AK 160
> Hs22041268
Length=155
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 40/66 (60%), Gaps = 8/66 (12%)
Query 83 KKFVIHIERISREKANGESL-IGIHPSKVVITKPKLNKDRK---ALLARKGKA----ANK 134
KK+VI+ ER+ EK+NG ++ GIHPS+ VI + L+ D K +L K K+ +
Sbjct 83 KKYVIYTERVQGEKSNGTTVHAGIHPSQAVIIRLTLDNDHKKIQQILEWKAKSHHVGNER 142
Query 135 GKYTEK 140
GKY E+
Sbjct 143 GKYKEE 148
> CE26135
Length=438
Score = 31.6 bits (70), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 43 CKELRKKFNVRSLPIRKDDEVMIVRGRFHDREGKV 77
C+EL F+V++ + D ++ + FHD G+V
Sbjct 234 CEELESGFSVKAYWVSTQDHLLNISSTFHDDSGRV 268
> Hs4557513
Length=500
Score = 28.5 bits (62), Expect = 4.2, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 53/97 (54%), Gaps = 15/97 (15%)
Query 52 VRSLPIRKDDEVMIVRGRFHDREGKVTQVYRKKFVIHIERISREKANGESLIGI--HPSK 109
VR L I+K DEV+ VR R H K Q F++ + +++ N ++L+ + H SK
Sbjct 47 VRDLTIQKADEVVWVRARVHTSRAKGKQC----FLV----LRQQQFNVQALVAVGDHASK 98
Query 110 VVITKPKLNKDRKALLARKG--KAANK--GKYTEKDV 142
++ K N ++++++ +G + N+ G T++DV
Sbjct 99 QMV-KFAANINKESIVDVEGVVRKVNQKIGSCTQQDV 134
> SPBC19C2.01
Length=739
Score = 27.7 bits (60), Expect = 7.2, Method: Composition-based stats.
Identities = 34/133 (25%), Positives = 58/133 (43%), Gaps = 21/133 (15%)
Query 25 AHFGAPSSVRRKIMTAP-----LCKELRKKFNVRSLPIRKDDEVMIVRGRFHDRE----- 74
+ F + RK+ P L ELRKK + L R ++ I+R D E
Sbjct 209 SKFSSEELALRKLADDPESWRKLASELRKKSRQQYLKPRAQQQLEILRREIRDEEQLFAG 268
Query 75 -----GKVTQVYRKKFVIHI--ERISREKANGESLIGIHPSKVVITKPKLNKDRK-ALLA 126
++ ++ +KK ++ I ER EK E + P + KL++ RK +L
Sbjct 269 EKLTQAEIRELEKKKELLRIAEERQRLEKQATEYQM---PEDYFTEQGKLDRKRKEEVLY 325
Query 127 RKGKAANKGKYTE 139
++ K +N+G+ E
Sbjct 326 QRYKDSNEGEQNE 338
Lambda K H
0.320 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40