bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1081_orf1
Length=336
Score E
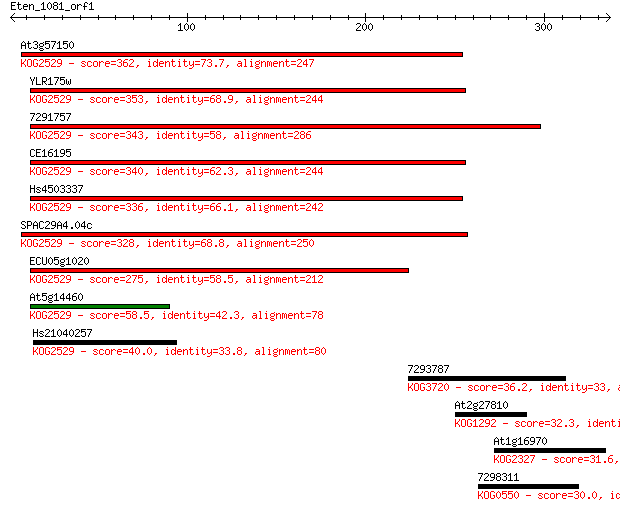
Sequences producing significant alignments: (Bits) Value
At3g57150 362 7e-100
YLR175w 353 4e-97
7291757 343 4e-94
CE16195 340 4e-93
Hs4503337 336 5e-92
SPAC29A4.04c 328 7e-90
ECU05g1020 275 1e-73
At5g14460 58.5 2e-08
Hs21040257 40.0 0.007
7293787 36.2 0.11
At2g27810 32.3 1.7
At1g16970 31.6 2.9
7298311 30.0 7.8
> At3g57150
Length=565
Score = 362 bits (929), Expect = 7e-100, Method: Compositional matrix adjust.
Identities = 182/248 (73%), Positives = 213/248 (85%), Gaps = 1/248 (0%)
Query 7 DVASM-QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGT 65
DVA + +ALE+LTGA+FQRPP+ISAVKRQLRIRTIYES++L YD D+HL VF VSCEAGT
Sbjct 156 DVAKVARALESLTGAVFQRPPLISAVKRQLRIRTIYESKLLEYDADRHLVVFWVSCEAGT 215
Query 66 YIRTLCVHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRR 125
YIRT+CVHLGLLLG G HMQELRRVRSG L E+ N+VT+HDV+DA FVYD RDE+YLRR
Sbjct 216 YIRTMCVHLGLLLGVGGHMQELRRVRSGILGENNNMVTMHDVMDAQFVYDNSRDESYLRR 275
Query 126 IIFPLEMLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIA 185
+I PLEM+L R+VVKDS VNAICYGAKLMIPG+LRFEN+I+V EVVLMTTKGEAIA
Sbjct 276 VIMPLEMILTSYKRLVVKDSAVNAICYGAKLMIPGLLRFENDIDVGTEVVLMTTKGEAIA 335
Query 186 LGIAQMSTPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRP 245
+GIA+M+T V+A+ DHG+VA +KRV+MDRDTY +WG GPRAS KKK+I G LDK G+P
Sbjct 336 VGIAEMTTSVMATCDHGVVAKIKRVVMDRDTYPRKWGLGPRASMKKKLIADGKLDKHGKP 395
Query 246 NENTPKEW 253
NE TP EW
Sbjct 396 NEKTPVEW 403
> YLR175w
Length=483
Score = 353 bits (905), Expect = 4e-97, Method: Compositional matrix adjust.
Identities = 168/244 (68%), Positives = 200/244 (81%), Gaps = 0/244 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
++LE LTGALFQRPP+ISAVKRQLR+RTIYES ++ +D ++L VF SCEAGTY+RTLC
Sbjct 141 RSLENLTGALFQRPPLISAVKRQLRVRTIYESNLIEFDNKRNLGVFWASCEAGTYMRTLC 200
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLG+LLG G HMQELRRVRSG L E+ N+VTLHDV+DA +VYD RDE+YLR II PLE
Sbjct 201 VHLGMLLGVGGHMQELRRVRSGALSENDNMVTLHDVMDAQWVYDNTRDESYLRSIIQPLE 260
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LLVG RIVVKDS VNA+CYGAKLMIPG+LR+E IE+ E+VL+TTKGEAIA+ IAQM
Sbjct 261 TLLVGYKRIVVKDSAVNAVCYGAKLMIPGLLRYEEGIELYDEIVLITTKGEAIAVAIAQM 320
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
ST +AS DHG+VA+VKR +M+RD Y RWG GP A +KK+M G LDK GR NENTP+
Sbjct 321 STVDLASCDHGVVASVKRCIMERDLYPRRWGLGPVAQKKKQMKADGKLDKYGRVNENTPE 380
Query 252 EWLK 255
+W K
Sbjct 381 QWKK 384
> 7291757
Length=508
Score = 343 bits (879), Expect = 4e-94, Method: Compositional matrix adjust.
Identities = 166/290 (57%), Positives = 215/290 (74%), Gaps = 6/290 (2%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
Q LE L GALFQRPP+ISAVKRQLR+RT+Y+S++L YDE +++ VF VSCEAG+YIRT+C
Sbjct 169 QGLEKLRGALFQRPPLISAVKRQLRVRTVYDSKLLDYDETRNMGVFWVSCEAGSYIRTMC 228
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGL+LG G M ELRRVRSG E +VT+HDVLDAM++Y+ +DE+ LRR+I PLE
Sbjct 229 VHLGLVLGVGGQMLELRRVRSGIQSERDGMVTMHDVLDAMWLYENHKDESMLRRVIKPLE 288
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LLV RI++KDS VNA+CYGAK+ +PGVLR+E+ IE++ E+V+ TTKGEAI L IA M
Sbjct 289 GLLVNHKRIIMKDSSVNAVCYGAKITLPGVLRYEDGIEIDQEIVICTTKGEAICLAIALM 348
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T +AS DHG+VA +KRV+M+RDTY +WG GP+AS KK +I G LDK GRPNENTPK
Sbjct 349 TTATMASCDHGVVAKIKRVIMERDTYPRKWGLGPKASAKKALIAAGKLDKFGRPNENTPK 408
Query 252 EWLKTEGFLPKLTGEPEA----PAENAVKSEREEGAPECEEGTAQNGVTE 297
EWL G++ +P A P + + + + + E TA V+E
Sbjct 409 EWL--TGYVDYNAKKPAAQEVSPTNGSSEPSKRKLSTSSVEETAAAAVSE 456
> CE16195
Length=445
Score = 340 bits (871), Expect = 4e-93, Method: Compositional matrix adjust.
Identities = 152/244 (62%), Positives = 196/244 (80%), Gaps = 0/244 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
QALE LTGALFQRPP+ISAVKRQLRIRT+YE++ + YD + + +F CE+GTY+RT+C
Sbjct 159 QALEKLTGALFQRPPLISAVKRQLRIRTVYENKFIEYDPAQQMGIFNCICESGTYVRTIC 218
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGL+LG G MQELRR RSG E+ N+VT+HDVLDA ++ D +DE+Y+R I+ PLE
Sbjct 219 VHLGLILGCGGQMQELRRNRSGICDENENMVTMHDVLDAQYLLDTQKDESYMRHIVRPLE 278
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL R+VVKDSC+NAICYGAK++IPG+LR++++IEV E+V+M+TKGEAI + IAQM
Sbjct 279 ALLTQHKRVVVKDSCINAICYGAKILIPGILRYDDDIEVGKEIVIMSTKGEAICIAIAQM 338
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T IASVDHG+VA KRV+M+RD Y +WG GP AS+KK+M+ GLLDK G+PN+ TPK
Sbjct 339 NTSTIASVDHGVVAKSKRVIMERDVYGRKWGLGPVASKKKQMVKDGLLDKFGKPNDTTPK 398
Query 252 EWLK 255
W K
Sbjct 399 SWAK 402
> Hs4503337
Length=514
Score = 336 bits (861), Expect = 5e-92, Method: Compositional matrix adjust.
Identities = 160/242 (66%), Positives = 203/242 (83%), Gaps = 0/242 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+ALETLTGALFQRPP+I+AVKRQLR+RTIYES+++ YD ++ L +F VSCEAGTYIRTLC
Sbjct 171 RALETLTGALFQRPPLIAAVKRQLRVRTIYESKMIEYDPERRLGIFWVSCEAGTYIRTLC 230
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
VHLGLLLG G MQELRRVRSG + E ++VT+HDVLDA ++YD +DE+YLRR+++PLE
Sbjct 231 VHLGLLLGVGGQMQELRRVRSGVMSEKDHMVTMHDVLDAQWLYDNHKDESYLRRVVYPLE 290
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LL R+V+KDS VNAICYGAK+M+PGVLR+E+ IEVN E+V++TTKGEAI + IA M
Sbjct 291 KLLTSHKRLVMKDSAVNAICYGAKIMLPGVLRYEDGIEVNQEIVVITTKGEAICMAIALM 350
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPNENTPK 251
+T VI++ DHGIVA +KRV+M+RDTY +WG GP+AS+KK MI GLLDK G+P ++TP
Sbjct 351 TTAVISTCDHGIVAKIKRVIMERDTYPRKWGLGPKASQKKLMIKQGLLDKHGKPTDSTPA 410
Query 252 EW 253
W
Sbjct 411 TW 412
> SPAC29A4.04c
Length=474
Score = 328 bits (842), Expect = 7e-90, Method: Compositional matrix adjust.
Identities = 172/250 (68%), Positives = 209/250 (83%), Gaps = 2/250 (0%)
Query 7 DVASMQALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTY 66
+VAS A+ETLTGALFQRPP+ISAVKRQLRIR+IYES+++ +D +++L VF SCEAGTY
Sbjct 143 NVAS--AIETLTGALFQRPPLISAVKRQLRIRSIYESKLIEFDNERNLAVFWASCEAGTY 200
Query 67 IRTLCVHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRI 126
+RTLCVHLGLLLG G HMQELRRVRSG L E+ ++VT+HDVLDA ++YD RDE+YLRR+
Sbjct 201 MRTLCVHLGLLLGVGGHMQELRRVRSGCLSENDDIVTMHDVLDAQWIYDNTRDESYLRRV 260
Query 127 IFPLEMLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIAL 186
I PLE LLVG RIVVKDS VNAICYGAKLMIPG+LR+E IEVN E+VL+TTKGEAIA+
Sbjct 261 IRPLESLLVGYKRIVVKDSAVNAICYGAKLMIPGLLRYEAGIEVNEEIVLITTKGEAIAV 320
Query 187 GIAQMSTPVIASVDHGIVAAVKRVLMDRDTYAMRWGYGPRASEKKKMILGGLLDKKGRPN 246
GIAQMST +++ DHG+VA VKR +M+RD Y RWG GP++ +KK + G LDK GRPN
Sbjct 321 GIAQMSTVELSTCDHGVVAKVKRCIMERDVYPRRWGLGPQSMKKKTLKKEGKLDKYGRPN 380
Query 247 ENTPKEWLKT 256
ENTP +W K+
Sbjct 381 ENTPADWSKS 390
> ECU05g1020
Length=357
Score = 275 bits (702), Expect = 1e-73, Method: Compositional matrix adjust.
Identities = 124/212 (58%), Positives = 170/212 (80%), Gaps = 1/212 (0%)
Query 12 QALETLTGALFQRPPVISAVKRQLRIRTIYESEILSYDEDKHLCVFRVSCEAGTYIRTLC 71
+A + L G+LFQRPP++ AVKR+LRIR +Y +IL ++ +K L +F+VSCEAGTY+RTLC
Sbjct 138 KASKRLVGSLFQRPPLMCAVKRELRIRNVYSIDILEFNREKRLGLFKVSCEAGTYVRTLC 197
Query 72 VHLGLLLGTGAHMQELRRVRSGNLQEDGNLVTLHDVLDAMFVYDAFRDETYLRRIIFPLE 131
H+G+L+G GA M+ELRR RSG + E + TLHD+LDA+++++ +DET +RR+I PLE
Sbjct 198 THIGILMGCGARMKELRRTRSGRISEK-DAFTLHDLLDAVYIFNTTKDETVIRRVILPLE 256
Query 132 MLLVGLPRIVVKDSCVNAICYGAKLMIPGVLRFENNIEVNAEVVLMTTKGEAIALGIAQM 191
LLVG PRI++KDSCVNAICYGAKL I GVLRF+ +I+V EVV+MTTKGEA+ALGIA +
Sbjct 257 SLLVGYPRIMIKDSCVNAICYGAKLSITGVLRFDRDIDVGTEVVIMTTKGEAVALGIALV 316
Query 192 STPVIASVDHGIVAAVKRVLMDRDTYAMRWGY 223
S+P ++ +DHG+V KRV+M++D Y WG+
Sbjct 317 SSPEMSIIDHGLVCRTKRVIMEKDLYPRSWGF 348
> At5g14460
Length=540
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 50/95 (52%), Gaps = 17/95 (17%)
Query 12 QALETLTGALFQRPPVISAVKR---------------QLRIR--TIYESEILSYDEDKHL 54
+AL + G ++Q PP+ SA+K +L R +I++ +I +D+
Sbjct 420 KALTSFLGEIWQVPPMFSAIKVGGEKMYEKARRGETVELSPRRISIFQFDIERSLDDRQN 479
Query 55 CVFRVSCEAGTYIRTLCVHLGLLLGTGAHMQELRR 89
+FRV C GTYIR+LC L LG+ AH+ LRR
Sbjct 480 LIFRVICSKGTYIRSLCADLAKALGSCAHLTALRR 514
> Hs21040257
Length=349
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 47/96 (48%), Gaps = 16/96 (16%)
Query 14 LETLTGALFQRPPVISAVKRQ-LRIRTIYE-SEILSYDEDKHLCVFRVS----------- 60
L+ TG + Q PP+ SA+K+ R+ T+ + E++ + + V+ +S
Sbjct 186 LQKFTGNIMQVPPLYSALKKDGQRLSTLMKRGEVVEAKPARPVTVYSISLQKFQPPFFTL 245
Query 61 ---CEAGTYIRTLCVHLGLLLGTGAHMQELRRVRSG 93
C G YIR+L +G L + A++ EL R + G
Sbjct 246 DVECGGGFYIRSLVSDIGKELSSCANVLELTRTKQG 281
> 7293787
Length=410
Score = 36.2 bits (82), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 11/89 (12%)
Query 224 GPRASEKKKMILGGLLDKKGRPNENTPKEWLKTEGFLP-KLTGEPEAPAENAVKSEREEG 282
PR +M+L GL P ENTP EW + + P + EPE E V +
Sbjct 131 SPRTLMSMQMVLAGLF-----PPENTPMEWNQLLNWQPIPIVMEPE---ETDVHIRMKAP 182
Query 283 APECEEGTAQNGVTEKPKKKKRHIEGEDV 311
P +E + V E P+ KK H E D+
Sbjct 183 CPRYDESVLE--VIELPEVKKLHAESSDL 209
> At2g27810
Length=721
Score = 32.3 bits (72), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 6/46 (13%)
Query 250 PKEWLKTEGFLPKLTGEPEAPAENA------VKSEREEGAPECEEG 289
P W K GF PK +GE A ++ V+++++E P+ E G
Sbjct 26 PSSWAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAG 71
> At1g16970
Length=610
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 272 ENAVKSEREEGAPECEEGTAQNGVTEKPKKKKRHIEGEDVEDEEQADKPSKKHKKKKHKD 331
EN ++ R+E P+ EEG + V + ++ K+ I G+D DEE +K KK+K D
Sbjct 500 ENELRETRDETLPD-EEGMNRPAVVKAIEQFKQSIYGDD-PDEESDSGAKEKSKKRKAGD 557
Query 332 AE 333
A+
Sbjct 558 AD 559
> 7298311
Length=508
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 263 LTGEPEAPAENAVKSEREEGAPECEEGTAQNGVTEKPKKKKRHIEGEDVEDEEQAD 318
L P+ A ++ + +EE E G A + KK R+ G+D+E++EQAD
Sbjct 427 LVHHPDRHANSSAEERKEEELKFKEVGEAY-AILSDAHKKSRYDSGQDIEEQEQAD 481
Lambda K H
0.317 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7988630762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40