bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1075_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
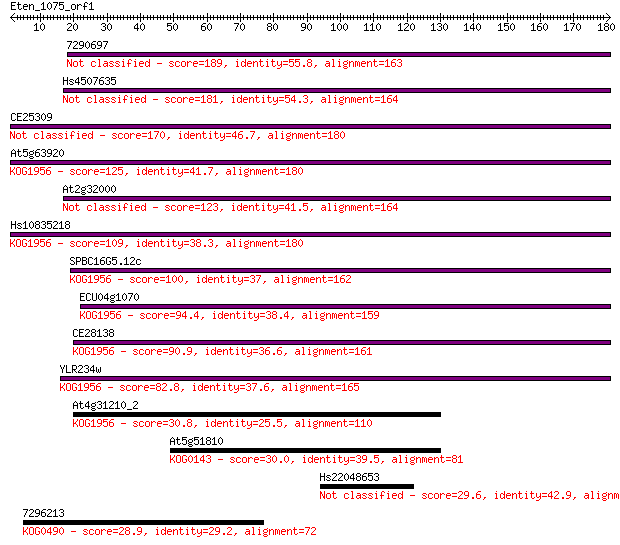
7290697 189 3e-48
Hs4507635 181 9e-46
CE25309 170 1e-42
At5g63920 125 6e-29
At2g32000 123 2e-28
Hs10835218 109 3e-24
SPBC16G5.12c 100 2e-21
ECU04g1070 94.4 1e-19
CE28138 90.9 1e-18
YLR234w 82.8 3e-16
At4g31210_2 30.8 1.6
At5g51810 30.0 2.5
Hs22048653 29.6 3.0
7296213 28.9 5.3
> 7290697
Length=875
Score = 189 bits (480), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 91/163 (55%), Positives = 117/163 (71%), Gaps = 2/163 (1%)
Query 18 QQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQGKY 77
Q +RAHFSA+ DI+KAM+ LG PN++E+ +VDARQELDL+IG AFTRFQT++FQ +Y
Sbjct 144 QVTYRAHFSAITEKDIKKAMETLGHPNENEAKSVDARQELDLRIGCAFTRFQTKFFQDRY 203
Query 78 GDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVGEQELTLEWARGQVF 137
GDLDSSLISYGPCQTPTL CV R D IQ F+PE ++ +Q + G+ E+TLEWARG+VF
Sbjct 204 GDLDSSLISYGPCQTPTLGFCVKRHDDIQTFKPESFWHLQLL--AGQPEVTLEWARGRVF 261
Query 138 DRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALLK 180
+ +A M V+ A V + E PQALNTV L++
Sbjct 262 KKDIAIMLLNRVKEHKKATVESVASKEAYKSKPQALNTVELMR 304
> Hs4507635
Length=862
Score = 181 bits (458), Expect = 9e-46, Method: Compositional matrix adjust.
Identities = 89/165 (53%), Positives = 117/165 (70%), Gaps = 1/165 (0%)
Query 17 QQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQGK 76
++ ++RA FS++ TDI AM LG P+ +E+ +VDARQELDL+IG AFTRFQT+YFQGK
Sbjct 144 EKTVFRARFSSITDTDICNAMACLGEPDHNEALSVDARQELDLRIGCAFTRFQTKYFQGK 203
Query 77 YGDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVG-EQELTLEWARGQ 135
YGDLDSSLIS+GPCQTPTL CV R D IQ+F+PE Y+ +Q V ++ L L+W R +
Sbjct 204 YGDLDSSLISFGPCQTPTLGFCVERHDKIQSFKPETYWVLQAKVNTDKDRSLLLDWDRVR 263
Query 136 VFDRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALLK 180
VFDR +AQMF + + A V T + E+ P ALNTV +L+
Sbjct 264 VFDREIAQMFLNMTKLEKEAQVEATSRKEKAKQRPLALNTVEMLR 308
> CE25309
Length=615
Score = 170 bits (431), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 84/182 (46%), Positives = 118/182 (64%), Gaps = 2/182 (1%)
Query 1 VLDLCKAHLVKPPNKQQQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLK 60
V+D K + K ++RAHFSA+ DI+ AM+ LG P+++ S +VDARQELDL+
Sbjct 129 VIDAVKCAMFKNRQDFMSDVYRAHFSAITEKDIKAAMRNLGRPDQNMSHSVDARQELDLR 188
Query 61 IGVAFTRFQTQYFQGKYGDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIV 120
IG +FTRFQT++FQGKYGDLDS++ISYGPCQTPTL CV R D I F+PE+Y+ ++
Sbjct 189 IGCSFTRFQTKFFQGKYGDLDSNVISYGPCQTPTLGFCVTRHDQIVQFKPEQYWVLKTNF 248
Query 121 RVGE--QELTLEWARGQVFDRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVAL 178
+ Q + EW RG++FD VA++F ++ +V + E P ALNTV L
Sbjct 249 TTSDDGQIFSPEWQRGRIFDAEVARVFVDRIKKCHTGLVLDVSKKEARKERPCALNTVEL 308
Query 179 LK 180
++
Sbjct 309 MR 310
> At5g63920
Length=926
Score = 125 bits (313), Expect = 6e-29, Method: Composition-based stats.
Identities = 75/186 (40%), Positives = 102/186 (54%), Gaps = 12/186 (6%)
Query 1 VLDLCKAHLVKPPNKQQQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLK 60
V+D+C+A K I RAHFSAL DI +A++ L PN+ ++AVDARQE+DL+
Sbjct 135 VVDVCRA------VKHNLFIRRAHFSALIDRDIHEAVQNLRDPNQLFAEAVDARQEIDLR 188
Query 61 IGVAFTRFQTQYFQGKY-----GDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQ 115
IG +FTRFQT + ++ G+ S +ISYGPCQ PTL V R IQA PEE++
Sbjct 189 IGASFTRFQTMLLRDRFAIDSTGEERSRVISYGPCQFPTLGFIVERYWEIQAHEPEEFWT 248
Query 116 IQPIVRVGEQELTLEWARGQVFDRVVAQ-MFHFIVQSATFAVVSTTKQTEEVLPMPQALN 174
I + E T W RG +FD A ++ V+ T V++ E P LN
Sbjct 249 INCSHQSEEGLATFNWMRGHLFDYASAVILYEMCVEEPTATVMNVPHPRERFKYPPYPLN 308
Query 175 TVALLK 180
T+ L K
Sbjct 309 TIELEK 314
> At2g32000
Length=836
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 68/164 (41%), Positives = 92/164 (56%), Gaps = 18/164 (10%)
Query 17 QQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQGK 76
+++++RA FS++ DI KAM L PN+DE+ AVDARQE+DLKI YF+
Sbjct 177 KRKVYRARFSSVTEKDISKAMDNLVEPNRDEALAVDARQEIDLKI------LLNVYFR-- 228
Query 77 YGDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVGEQELTLEWARGQV 136
YGPCQTPTL CV R I F+PE+++ ++P +R EL LEW R ++
Sbjct 229 ----------YGPCQTPTLGFCVQRYMHINTFKPEKFWALRPYIRKDGYELQLEWERRRL 278
Query 137 FDRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALLK 180
FD A +F +V A V + +EV P LNTV LLK
Sbjct 279 FDLEAATVFQKLVVEGRTAKVMDVSEKQEVKGRPAGLNTVNLLK 322
> Hs10835218
Length=1001
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 69/181 (38%), Positives = 97/181 (53%), Gaps = 7/181 (3%)
Query 1 VLDLCKAHLVKPPNKQQQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLK 60
++ +CKA VKP Q+ RA FS + P ++ A + L P++ SDAVD RQELDL+
Sbjct 160 IIHVCKA--VKP----NLQVLRARFSEITPHAVRTACENLTEPDQRVSDAVDVRQELDLR 213
Query 61 IGVAFTRFQTQYFQGKYGD-LDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPI 119
IG AFTRFQT Q + + L LISYG CQ PTL V R IQAF PE +++I+
Sbjct 214 IGAAFTRFQTLRLQRIFPEVLAEQLISYGSCQFPTLGFVVERFKAIQAFVPEIFHRIKVT 273
Query 120 VRVGEQELTLEWARGQVFDRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALL 179
+ + W R ++F+ + + + A V + + PQAL+TV L
Sbjct 274 HDHKDGIVEFNWKRHRLFNHTACLVLYQLCVEDPMATVVEVRSKPKSKWRPQALDTVELE 333
Query 180 K 180
K
Sbjct 334 K 334
> SPBC16G5.12c
Length=622
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/164 (36%), Positives = 90/164 (54%), Gaps = 4/164 (2%)
Query 19 QIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQGKYG 78
Q+ RA F+ L + I A K+ +K+ +DAVDAR ELD ++G FTR QT Q +
Sbjct 141 QVIRADFNNLERSHIISAAKRPRDVSKNAADAVDARIELDFRLGAIFTRLQTIQLQKSFD 200
Query 79 DLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVGEQELTLEWARGQVFD 138
L + +ISYGPCQ PTL V R ++ F PE Y+ ++ + + + + W R +VFD
Sbjct 201 ILQNKIISYGPCQFPTLGFVVDRWQRVEDFVPETYWHLRFVDKRQGKTIQFNWERAKVFD 260
Query 139 RVVAQ-MFHFIVQSATFAVVSTT-KQTEEVLPMPQALNTVALLK 180
R+ + ++ T VV+ T K + P+P L+TV L K
Sbjct 261 RLTTMIILENCLECKTAKVVNITQKPKTKYKPLP--LSTVELTK 302
> ECU04g1070
Length=621
Score = 94.4 bits (233), Expect = 1e-19, Method: Composition-based stats.
Identities = 61/166 (36%), Positives = 89/166 (53%), Gaps = 17/166 (10%)
Query 22 RAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYF----QGKY 77
RA FS + D++ A++ LG N+ E++AV+ R ELDL+IG AFTR QT QGK
Sbjct 33 RARFSGITLRDVESALRNLGEINEAEAEAVEVRIELDLRIGSAFTRLQTLSLGRARQGK- 91
Query 78 GDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYY--QIQPIVRVGEQELTLEWARGQ 135
++S+GPCQ PTL V R+ ++F PE ++ + + + + + E T W RG
Sbjct 92 -----KILSFGPCQVPTLGFVVERARERESFVPEIFWTLKFKAMHKGFKDEFT--WRRGP 144
Query 136 VFDR-VVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALLK 180
VFDR V F+ + V K+ + P+P L TV L K
Sbjct 145 VFDRNCVVCFFNSLSGEDARIVSREVKEKTKYRPLP--LRTVELQK 188
> CE28138
Length=759
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 59/168 (35%), Positives = 85/168 (50%), Gaps = 9/168 (5%)
Query 20 IWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQGKYGD 79
I+RA FS + I +A + L ++ AVD R ELDL+IG AFTR QT + + ++ D
Sbjct 141 IFRARFSEITKAAITRAARNLIRLDEKTVAAVDCRSELDLRIGSAFTRLQTLHLRNRFRD 200
Query 80 L-----DSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVGEQELTLE--WA 132
L S +ISYG CQ PTL R I+ F E ++++ IV + +E W
Sbjct 201 LLGQNDTSQVISYGSCQFPTLGFVTDRYKMIENFVSEPFWKL--IVEHTRESHKVEFLWD 258
Query 133 RGQVFDRVVAQMFHFIVQSATFAVVSTTKQTEEVLPMPQALNTVALLK 180
R ++FDR + H + A V + + PQAL+TV L K
Sbjct 259 RNRLFDRDTVDILHDECKETKEAHVEKVAKKPKSKWRPQALDTVELEK 306
> YLR234w
Length=656
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 62/187 (33%), Positives = 87/187 (46%), Gaps = 24/187 (12%)
Query 16 QQQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVAFTRFQTQYFQG 75
Q Q++RA FS L I A + + AV R E+DL+ GV FTR T+ +
Sbjct 144 QNDQVYRAVFSHLERQHILNAARNPSRLDMKSVHAVGTRIEIDLRAGVTFTRLLTETLRN 203
Query 76 KYGD----------------LDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPI 119
K + DS ++SYG CQ PTL V R + I+ F PEE++ IQ +
Sbjct 204 KLRNQATMTKDGAKHRGGNKNDSQVVSYGTCQFPTLGFVVDRFERIRNFVPEEFWYIQLV 263
Query 120 VRVGEQ--ELTLEWARGQVFDRVVAQMFH--FIVQSATFAVVS--TTKQTEEVLPMPQAL 173
V + T +W RG +FDR+ F+ I + A V +K T + P+P L
Sbjct 264 VENKDNGGTTTFQWDRGHLFDRLSVLTFYETCIETAGNVAQVVDLKSKPTTKYRPLP--L 321
Query 174 NTVALLK 180
TV L K
Sbjct 322 TTVELQK 328
> At4g31210_2
Length=676
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 46/113 (40%), Gaps = 14/113 (12%)
Query 20 IWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDA---RQELDLKIGVAFTRFQTQYFQGK 76
+ R F + + I+ A++ SP + + D V A R+ LD IG + + G
Sbjct 45 VARVVFHEITESAIKSALQ---SPREIDGDLVHAYLARRALDYLIGFNISPLLWRKLPG- 100
Query 77 YGDLDSSLISYGPCQTPTLQLCVHRSDTIQAFRPEEYYQIQPIVRVGEQELTL 129
S G Q+ L L R I F+P+EY+ + V+ + T
Sbjct 101 -------CPSAGRVQSAALALVCDRESEIDGFKPQEYWTVGIKVKGKDNSATF 146
> At5g51810
Length=378
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 45/106 (42%), Gaps = 26/106 (24%)
Query 49 DAVDARQELDLKI----GVAFTRFQTQYFQGKYGDLDS--SLISYGPCQTPTLQL----- 97
D +A L LKI G++ YF+G + + DS L Y PCQTP L L
Sbjct 187 DYCEAMSSLSLKIMELLGLSLG-VNRDYFRGFFEENDSIMRLNHYPPCQTPDLTLGTGPH 245
Query 98 CVHRSDTI---------QAFRPEEYYQIQP-----IVRVGEQELTL 129
C S TI Q F ++ I+P +V +G+ + L
Sbjct 246 CDPSSLTILHQDHVNGLQVFVDNQWQSIRPNPKAFVVNIGDTFMAL 291
> Hs22048653
Length=485
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 94 TLQLCVHRSDTIQAFRPEEYYQIQPIVR 121
+ Q C+H DT Q F E+Y I P +R
Sbjct 25 SFQFCMHVQDTSQLFGGAEFYSIDPSLR 52
> 7296213
Length=408
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query 5 CKAHLVKPPNKQQQQIWRAHFSALAPTDIQKAMKQLGSPNKDESDAVDARQELDLKIGVA 64
C+A P K++Q+ +R F++ +++KA + P+ V R+EL +KIG+
Sbjct 74 CEADEYAP--KRKQRRYRTTFTSFQLEELEKAFSRTHYPD------VFTREELAMKIGLT 125
Query 65 FTRFQTQYFQGK 76
R Q +FQ +
Sbjct 126 EARIQV-WFQNR 136
Lambda K H
0.320 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40