bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1074_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
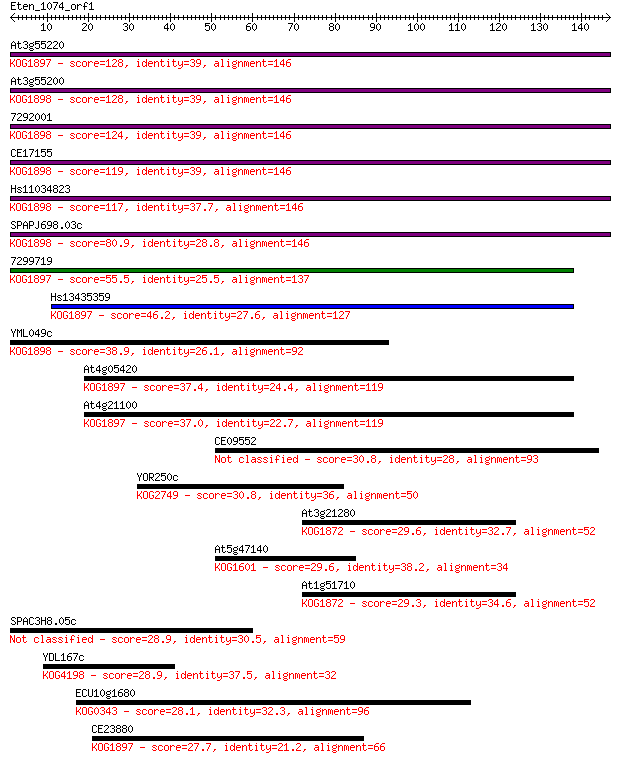
At3g55220 128 4e-30
At3g55200 128 4e-30
7292001 124 7e-29
CE17155 119 2e-27
Hs11034823 117 8e-27
SPAPJ698.03c 80.9 7e-16
7299719 55.5 3e-08
Hs13435359 46.2 2e-05
YML049c 38.9 0.003
At4g05420 37.4 0.009
At4g21100 37.0 0.012
CE09552 30.8 0.93
YOR250c 30.8 1.1
At3g21280 29.6 1.9
At5g47140 29.6 2.2
At1g51710 29.3 2.5
SPAC3H8.05c 28.9 4.0
YDL167c 28.9 4.1
ECU10g1680 28.1 5.5
CE23880 27.7 7.1
> At3g55220
Length=1214
Score = 128 bits (321), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 94/146 (64%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
GKKRLL+KCE K P+ I+ ++ DRI+ GD+ ES H KY+ + N Y+ DD PRW
Sbjct 971 GKKRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHYCKYRRDENQLYIFADDCVPRW 1030
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
LT +D+ ++ ADKF +++ R+P + ++ D TG K++ + + + +K+D +
Sbjct 1031 LTASHHVDFDTMAGADKFGNVYFVRLPQDLSEEIEEDPTGGKIKWEQGKLNGAPNKVDEI 1090
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
+QFHVG+ VT L++A++ G SE+I+
Sbjct 1091 VQFHVGDVVTCLQKASMIPGGSESIM 1116
> At3g55200
Length=1214
Score = 128 bits (321), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 94/146 (64%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
GKKRLL+KCE K P+ I+ ++ DRI+ GD+ ES H KY+ + N Y+ DD PRW
Sbjct 971 GKKRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHYCKYRRDENQLYIFADDCVPRW 1030
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
LT +D+ ++ ADKF +++ R+P + ++ D TG K++ + + + +K+D +
Sbjct 1031 LTASHHVDFDTMAGADKFGNVYFVRLPQDLSEEIEEDPTGGKIKWEQGKLNGAPNKVDEI 1090
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
+QFHVG+ VT L++A++ G SE+I+
Sbjct 1091 VQFHVGDVVTCLQKASMIPGGSESIM 1116
> 7292001
Length=1169
Score = 124 bits (311), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 88/146 (60%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
GKK++L+KCE K++P IV ++ G R++ D+ ES+ I+Y+ N + DDT PRW
Sbjct 983 GKKKMLRKCENKHIPYQIVNIQAMGHRVYVSDVQESVFFIRYRRAENQLIIFADDTHPRW 1042
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
+T +LDY +I ADKF ++ I+R+P D D TG K D ++ + K + +
Sbjct 1043 VTATTLLDYDTIAIADKFGNLSIQRLPHSVTDDVDEDPTGTKSLWDRGLLSGASQKSENI 1102
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
FHVGE + L++ATL G SEA++
Sbjct 1103 CSFHVGEIIMSLQKATLIPGGSEALI 1128
> CE17155
Length=1220
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 87/146 (59%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
G+K+LL KCE KN P IV ++ G RI D ES+H ++Y+ N V DDT PR+
Sbjct 976 GQKKLLAKCENKNFPVSIVNIQSTGQRIIVSDSQESVHFLRYRKGDNQLVVFADDTTPRY 1035
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
+T VLDYH++ ADKF ++ + R+P +D D T K D ++ + K++ V
Sbjct 1036 VTCVCVLDYHTVAVADKFGNLAVVRLPERVNEDVQDDPTVSKSVWDRGWLNGASQKVELV 1095
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
F +G+T+T L++ +L GA+EA+V
Sbjct 1096 SNFFIGDTITSLQKTSLMPGANEALV 1121
> Hs11034823
Length=1217
Score = 117 bits (293), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 55/146 (37%), Positives = 85/146 (58%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
GKK+LL+KCE K++ + I ++ G R+ D+ ES ++YK N N + DDT PRW
Sbjct 973 GKKKLLRKCENKHIANYISGIQTIGHRVIVSDVQESFIWVRYKRNENQLIIFADDTYPRW 1032
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
+T +LDY ++ ADKF +I + R+P + D TG K D + + K + +
Sbjct 1033 VTTASLLDYDTVAGADKFGNICVVRLPPNTNDEVDEDPTGNKALWDRGLLNGASQKAEVI 1092
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
+ +HVGETV L++ TL G SE++V
Sbjct 1093 MNYHVGETVLSLQKTTLIPGGSESLV 1118
> SPAPJ698.03c
Length=1206
Score = 80.9 bits (198), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 70/146 (47%), Gaps = 0/146 (0%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
G K++L+K E +P I + V RI D S+ + YK N DDT RW
Sbjct 963 GNKKMLRKGELSAVPLFITHITVQASRIVVADSQYSVRFVVYKPEDNHLLTFADDTIHRW 1022
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
T ++DY ++ DKF +I++ R P + +++ KL + ++ + HKLD +
Sbjct 1023 TTTNVLVDYDTLAGGDKFGNIWLLRCPEHVSKLADEENSESKLIHEKPFLNSTPHKLDLM 1082
Query 121 LQFHVGETVTGLRRATLSTGASEAIV 146
F + T L++ L GA E ++
Sbjct 1083 AHFFTNDIPTSLQKVQLVEGAREVLL 1108
> 7299719
Length=1140
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 64/137 (46%), Gaps = 19/137 (13%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRW 60
+K L +C +N+ + + FLK GD I GD+ SI ++++K +F + D P+W
Sbjct 896 NEKELRMECNIQNMIAAL-FLKAKGDFILVGDLMRSITLLQHKQMEGIFVEIARDCEPKW 954
Query 61 LTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTV 120
+ E+LD + + ++ ++F+ + S A DE L +
Sbjct 955 MRAVEILDDDTFLGSETNGNLFVCQKDSAATTDE------------------ERQLLPEL 996
Query 121 LQFHVGETVTGLRRATL 137
+FH+G+TV R +L
Sbjct 997 ARFHLGDTVNVFRHGSL 1013
> Hs13435359
Length=1140
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 64/130 (49%), Gaps = 9/130 (6%)
Query 11 YKNLPSGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRWLTRGEVLDYH 70
Y N+ + ++LK GD I GD+ S+ ++ YK F + D P W++ E+LD
Sbjct 906 YNNIMA--LYLKTKGDFILVGDLMRSVLLLAYKPMEGNFEEIARDFNPNWMSAVEILDDD 963
Query 71 SIVAADKFDSIFIERVPSEARQDEGG---DSTGLKLRGDTAYMTDSCHKLDTVLQFHVGE 127
+ + A+ ++F+ + S A DE GL G+ ++ CH +++ ++GE
Sbjct 964 NFLGAENAFNLFVCQKDSAATTDEERQHLQEVGLFHLGE--FVNVFCH--GSLVMQNLGE 1019
Query 128 TVTGLRRATL 137
T T + + L
Sbjct 1020 TSTPTQGSVL 1029
> YML049c
Length=1361
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 49/98 (50%), Gaps = 9/98 (9%)
Query 1 GKKRLLKKCEYKNLPSGIVFLKVCG------DRIFAGDMSESIHVIKYKANANLFYVLCD 54
GKK+LL++ + + + K+ +R+ GD+ ES+ + + N+F D
Sbjct 1055 GKKQLLRRSVTQ---TPVSITKIVSMHQWNYERLAVGDIHESVTLFIWDPAGNVFIPYVD 1111
Query 55 DTGPRWLTRGEVLDYHSIVAADKFDSIFIERVPSEARQ 92
D+ R +T + LD +++ AD++ + + R P E +
Sbjct 1112 DSVKRHVTVLKFLDEATVIGADRYGNAWTLRSPPECEK 1149
> At4g05420
Length=1088
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 51/119 (42%), Gaps = 18/119 (15%)
Query 19 VFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRWLTRGEVLDYHSIVAADKF 78
++++ GD I GD+ +SI ++ YK D W++ E+LD + A+
Sbjct 871 LYVQTRGDFIVVGDLMKSISLLLYKHEEGAIEERARDYNANWMSAVEILDDDIYLGAENN 930
Query 79 DSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTVLQFHVGETVTGLRRATL 137
++ + SE DE RG +L+ V ++H+GE V R +L
Sbjct 931 FNLLTVKKNSEGATDEE--------RG----------RLEVVGEYHLGEFVNRFRHGSL 971
> At4g21100
Length=1088
Score = 37.0 bits (84), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 27/119 (22%), Positives = 49/119 (41%), Gaps = 18/119 (15%)
Query 19 VFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRWLTRGEVLDYHSIVAADKF 78
++++ GD I GD+ +SI ++ YK D W+T E+L+ + D
Sbjct 871 LYVQTRGDFIAVGDLMKSISLLIYKHEEGAIEERARDYNANWMTAVEILNDDIYLGTDNC 930
Query 79 DSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTVLQFHVGETVTGLRRATL 137
+IF + +E DE +++ V ++H+GE V R +L
Sbjct 931 FNIFTVKKNNEGATDE------------------ERARMEVVGEYHIGEFVNRFRHGSL 971
> CE09552
Length=1721
Score = 30.8 bits (68), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 42/95 (44%), Gaps = 7/95 (7%)
Query 51 VLCDDTGPRWLTRGEVLDYHSIVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYM 110
+LC+ P ++ +L V + +F E+VP +R D+ L+ T +
Sbjct 523 LLCEARMPTYVKMTAIL--RESVNSPEFSH---EKVPKLSRMHWNSDTLPAVLQVMTVSL 577
Query 111 TDSCHKLDTVLQFHVGETVTGL--RRATLSTGASE 143
+DS K D FHVG +V R + GA+E
Sbjct 578 SDSHSKEDCYHGFHVGMSVLNWVYRNVFIVRGAAE 612
> YOR250c
Length=445
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Query 32 DMSESIHVIKYKANANLFYVLCDDTGPRW----LTRGEVLDYHSIVAADKFDSI 81
+++E H+I+ K N N+ VLC +T P W T G L ++I K D +
Sbjct 261 NLAELHHIIE-KLNVNIMLVLCSETDPLWEKVKKTFGPELGNNNIFFIPKLDGV 313
> At3g21280
Length=488
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 72 IVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTVLQF 123
IV A + +F+E +P E + G S GL G+T YM + L +V +
Sbjct 77 IVKAPEKGPVFMEDLPEEQQAANLGYSAGLVNLGNTCYMNSTMQCLISVPEL 128
> At5g47140
Length=461
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 51 VLCDDTGPRWLTRGEVLDYHSIVAADKFDSIFIE 84
VLC+ G RW T+G +++Y + A + D IE
Sbjct 18 VLCNACGSRWRTKGSLVNYTPLHARAEGDETEIE 51
> At1g51710
Length=490
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 72 IVAADKFDSIFIERVPSEARQDEGGDSTGLKLRGDTAYMTDSCHKLDTVLQF 123
IV A + +F E +P EA G S GL G+T YM + L +V +
Sbjct 77 IVKAPEKAIVFAEDLPEEALATNLGYSAGLVNLGNTCYMNSTVQCLKSVPEL 128
> SPAC3H8.05c
Length=1073
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 5/61 (8%)
Query 1 GKKRLLKKCEYKNLP--SGIVFLKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGP 58
KK CE LP S IV + D ++ + +S+ V +Y + N ++C DT
Sbjct 909 SKKFCNSDCE---LPVRSPIVSMSTYKDYVYTSSLRDSVAVFQYDSENNSLNLVCSDTSS 965
Query 59 R 59
R
Sbjct 966 R 966
> YDL167c
Length=719
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 9 CEYKNLPSGIVFLKVCGDRIFAGDMSESIHVI 40
C Y N +V L+ G + +GD SE+ H I
Sbjct 590 CTYHNFAKNVVCLRCGGPKSISGDASETNHYI 621
> ECU10g1680
Length=452
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 45/120 (37%), Gaps = 30/120 (25%)
Query 17 GIVFLKVCGD---------------RIF--AGDMSES--IHVIKYKANANLFYVLCDDTG 57
GIVF C + RIF +G MS+ I V K + C D G
Sbjct 247 GIVFFSTCKEVKFHCLLFERLKLRNRIFCLSGGMSQKQRIDVFKRFVKEKNGILFCTDLG 306
Query 58 PRWLTRGEVLDYHSIVAADKFD-----SIFIERVPSEARQDEGGDSTGLKLRGDTAYMTD 112
R L D+ + ++D ++ RV AR E G+S + G+ +TD
Sbjct 307 SRGL------DFPKVDVVIQYDCPCNVETYVHRVGRTARNSERGESYVYLVHGEEKLLTD 360
> CE23880
Length=1134
Score = 27.7 bits (60), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/66 (21%), Positives = 29/66 (43%), Gaps = 0/66 (0%)
Query 21 LKVCGDRIFAGDMSESIHVIKYKANANLFYVLCDDTGPRWLTRGEVLDYHSIVAADKFDS 80
LKV + + D+ S+ ++ Y+ F + D +W+ E + SI+ + +
Sbjct 909 LKVMNEEVAVADVMRSVSLLSYRMLEGNFEEVAKDWNSQWMVTCEFITAESILGGEAHLN 968
Query 81 IFIERV 86
+F V
Sbjct 969 LFTVEV 974
Lambda K H
0.320 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40