bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1061_orf1
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
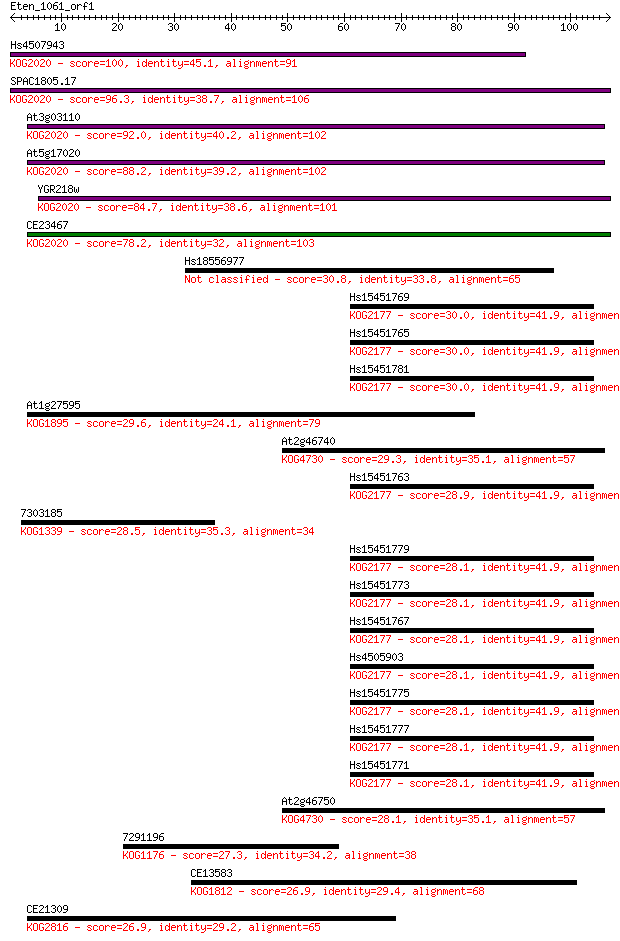
Hs4507943 100 8e-22
SPAC1805.17 96.3 1e-20
At3g03110 92.0 2e-19
At5g17020 88.2 3e-18
YGR218w 84.7 4e-17
CE23467 78.2 3e-15
Hs18556977 30.8 0.69
Hs15451769 30.0 1.1
Hs15451765 30.0 1.1
Hs15451781 30.0 1.2
At1g27595 29.6 1.3
At2g46740 29.3 1.9
Hs15451763 28.9 2.6
7303185 28.5 3.0
Hs15451779 28.1 3.8
Hs15451773 28.1 3.8
Hs15451767 28.1 3.8
Hs4505903 28.1 3.8
Hs15451775 28.1 3.9
Hs15451777 28.1 4.0
Hs15451771 28.1 4.1
At2g46750 28.1 4.6
7291196 27.3 7.8
CE13583 26.9 9.2
CE21309 26.9 9.9
> Hs4507943
Length=1071
Score = 100 bits (248), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 41/91 (45%), Positives = 61/91 (67%), Gaps = 0/91 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T+ +P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P TQ K +
Sbjct 816 TAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQAVNSHCFPAFLAIPPTQFKLVL 875
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
+S++WAFKH ++A+ GLQ+ LQ + +
Sbjct 876 DSIIWAFKHTMRNVADTGLQILFTLLQNVAQ 906
> SPAC1805.17
Length=966
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 61/106 (57%), Gaps = 0/106 (0%)
Query 1 TSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFV 60
T +P++ D +F CTL+MI DF YP+HR F+ LL+A N +CF L ++P+ Q K +
Sbjct 817 TDKIPLVLDAVFGCTLEMISKDFSEYPEHRAAFFQLLRAINLNCFPALLNIPAPQFKLVI 876
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYF 106
S+VW+FKH + E GL + E + + +V N F YY
Sbjct 877 NSIVWSFKHVSRDIQETGLNILLELINNMASMGPDVSNAFFQTYYI 922
> At3g03110
Length=1022
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 41/102 (40%), Positives = 65/102 (63%), Gaps = 6/102 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P+IF+ +F CTL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S+
Sbjct 769 VPLIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIATFCFRALIQLSSEQLKLVMDSV 828
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYY 105
+WAF+H ++AE GL + E L+ + +DFC +Y
Sbjct 829 IWAFRHTERNIAETGLNLLLEMLKNFQK------SDFCNKFY 864
> At5g17020
Length=1075
Score = 88.2 bits (217), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 64/102 (62%), Gaps = 6/102 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P IF+ +F CTL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S+
Sbjct 822 VPHIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIATFCFPALIKLSSPQLKLVMDSI 881
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYY 105
+WAF+H ++AE GL + E L+ + ++FC +Y
Sbjct 882 IWAFRHTERNIAETGLNLLLEMLKNFQQ------SEFCNQFY 917
> YGR218w
Length=1084
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 57/102 (55%), Gaps = 1/102 (0%)
Query 6 VIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVW 65
+I +F+CTL MI DF YP+HR FY LLK N+ F+ LP K FV+++ W
Sbjct 832 LILQSVFECTLDMINKDFTEYPEHRVEFYKLLKVINEKSFAAFLELPPAAFKLFVDAICW 891
Query 66 AFKHEHPSLAEEGLQVTHEFLQRLIE-GKREVLNDFCCNYYF 106
AFKH + + GLQ+ + ++ + G N+F NY+F
Sbjct 892 AFKHNNRDVEVNGLQIALDLVKNIERMGNVPFANEFHKNYFF 933
> CE23467
Length=1080
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 56/103 (54%), Gaps = 0/103 (0%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P I +F C++ MI D +++P+HR F+ L+ + Q CF +P L ++++
Sbjct 829 VPSILSAVFQCSIDMINKDMEAFPEHRTNFFELVLSLVQECFPVFMEMPPEDLGTVIDAV 888
Query 64 VWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYF 106
VWAF+H ++AE GL + E L R+ E ++ F YY
Sbjct 889 VWAFQHTMRNVAEIGLDILKELLARVSEQDDKIAQPFYKRYYI 931
> Hs18556977
Length=231
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 29/65 (44%), Gaps = 6/65 (9%)
Query 32 RFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE 91
RF LLKA++QH +G L+ W EHP L EGL E QR I
Sbjct 54 RFKPLLKASSQHVETGK----RRNLRETTAHSPWNENQEHPWLGVEGL--PKEKHQRKIN 107
Query 92 GKREV 96
++ +
Sbjct 108 PQQPI 112
> Hs15451769
Length=824
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451765
Length=829
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451781
Length=781
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> At1g27595
Length=1091
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 19/79 (24%), Positives = 30/79 (37%), Gaps = 4/79 (5%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
P + D++ + ++++ P F + H F L LP QL ES+
Sbjct 977 FPTLVDFVMEILSKLVRKQIWRLPKLWPGFLKCVSQTKPHSFPVLLELPVPQL----ESI 1032
Query 64 VWAFKHEHPSLAEEGLQVT 82
+ F PSL Q T
Sbjct 1033 MKKFPDLRPSLTAYANQPT 1051
> At2g46740
Length=590
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 8/63 (12%)
Query 49 FSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE------GKREVLNDFCC 102
FS+P TQ+K+F+ + K + SL GL++ + L R + GK DF
Sbjct 403 FSIPLTQVKSFINDIKSLLKIDSKSLC--GLELYYGILMRYVTSSPAYLGKETEAIDFDI 460
Query 103 NYY 105
YY
Sbjct 461 TYY 463
> Hs15451763
Length=882
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> 7303185
Length=404
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 3 VLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYAL 36
+LPV F + C LQ+ + + +P R RF L
Sbjct 13 ILPVQFQHPVSCKLQLYRVPLRRFPSARHRFEKL 46
> Hs15451779
Length=585
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451773
Length=611
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451767
Length=611
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs4505903
Length=633
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451775
Length=423
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451777
Length=435
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> Hs15451771
Length=560
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 61 ESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCN 103
E+ W KHE LAE Q EFL +G R+ N FC N
Sbjct 153 EAHQWFLKHEARPLAELRNQSVREFL----DGTRKTNNIFCSN 191
> At2g46750
Length=570
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 8/63 (12%)
Query 49 FSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIE------GKREVLNDFCC 102
S+P TQ+K+F+ + K E SL GL++ + L R + GK DF
Sbjct 378 LSVPLTQVKSFISDIKSLVKIEQKSLC--GLELHYGILMRYVTSSPAYLGKETEALDFDI 435
Query 103 NYY 105
YY
Sbjct 436 TYY 438
> 7291196
Length=543
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 21 SDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKA 58
++FQ YP+ E F L ++ C G+ ST L A
Sbjct 438 NNFQIYPEQIEEFILRLPGVSEACVFGIPDAVSTNLTA 475
> CE13583
Length=488
Score = 26.9 bits (58), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 27/68 (39%), Gaps = 4/68 (5%)
Query 33 FYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVTHEFLQRLIEG 92
F + N CF G F QL+ VE L WA E G H + R
Sbjct 423 FLSATPRKNLVCFLGKFEYQQAQLEKEVEELAWAV--ERADGTARGALEAH--MHRAEHK 478
Query 93 KREVLNDF 100
++ +L+DF
Sbjct 479 RQTLLHDF 486
> CE21309
Length=452
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 4 LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESL 63
+P+I YI C L I FQSY H ++ L+ A G ++ ST L V++
Sbjct 80 IPLIIPYI-GCILGTINYVFQSYFIHTSVYFLLISDALFGLCGGFIAIISTTLTYGVKTS 138
Query 64 VWAFK 68
+ ++
Sbjct 139 MLRYR 143
Lambda K H
0.328 0.140 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167556980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40