bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1039_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
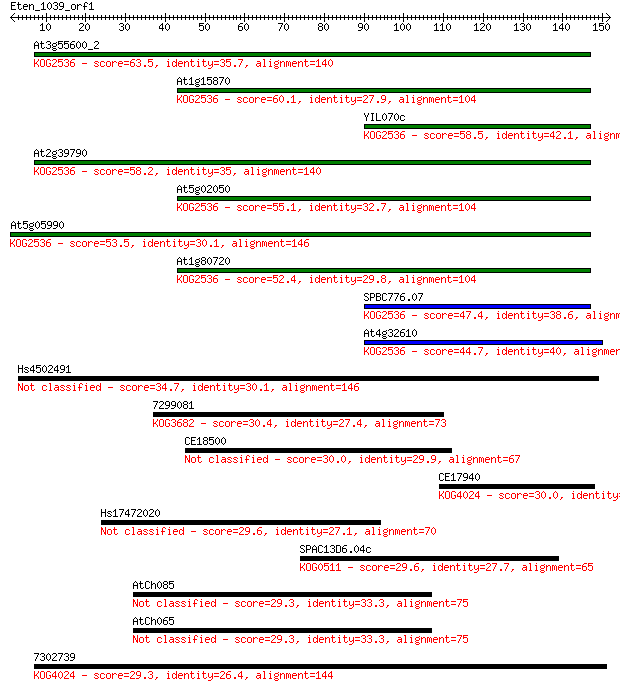
At3g55600_2 63.5 1e-10
At1g15870 60.1 2e-09
YIL070c 58.5 5e-09
At2g39790 58.2 6e-09
At5g02050 55.1 5e-08
At5g05990 53.5 1e-07
At1g80720 52.4 3e-07
SPBC776.07 47.4 1e-05
At4g32610 44.7 6e-05
Hs4502491 34.7 0.063
7299081 30.4 1.3
CE18500 30.0 1.7
CE17940 30.0 2.0
Hs17472020 29.6 2.2
SPAC13D6.04c 29.6 2.5
AtCh085 29.3 3.1
AtCh065 29.3 3.1
7302739 29.3 3.3
> At3g55600_2
Length=248
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 50/155 (32%), Positives = 70/155 (45%), Gaps = 21/155 (13%)
Query 7 VTLTREVDGLKVIVEFSCIQPAAA---------------AEEEAALPESSDFTVSVHRSD 51
VTLTRE +G ++ VE S P A E + S V+V +
Sbjct 98 VTLTREYNGEQIKVEVSM--PGLAMDENEDDVDDDEDGDGRHEKSNESSIPLVVTVTKKS 155
Query 52 GSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESLASYPGPYFEDLDEAFQQQLDSWLA 111
G L F C T DE G + D SS+E L +Y GP F++LDE ++ +L
Sbjct 156 GLSLEFSC-TAFPDEIV--IDGLSVNRPDDSSEEQL-TYDGPDFQELDENMRKSFHKFLE 211
Query 112 AVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
G+ DFL + K++REYL WL K+ F+
Sbjct 212 TRGIKASATDFLYEYMMKKDSREYLLWLKKLKTFV 246
> At1g15870
Length=190
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 53/104 (50%), Gaps = 6/104 (5%)
Query 43 FTVSVHRSDGSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESLASYPGPYFEDLDEAF 102
F V++ + DG L CS + I + FV SS+ S +Y GP F+++++
Sbjct 90 FIVNISKGDGETLEIMCSAWPD------TIQITKFFVRKSSQNSPNAYIGPEFQEMEDEL 143
Query 103 QQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
Q + +L G+ L +FL + +K+ EY+RW+ V +++
Sbjct 144 QDSVYRFLEERGISDDLAEFLHQYMKNKDKAEYIRWMETVKSYV 187
> YIL070c
Length=266
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 90 YPGPYFEDLDEAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
Y GP F +LDE Q+ L+++L + GV+ +L F+ A++ KEN EY+ WL K+ F
Sbjct 209 YHGPPFSNLDEELQESLEAYLESRGVNEELASFISAYSEFKENNEYISWLEKMKKFF 265
> At2g39790
Length=429
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 49/147 (33%), Positives = 67/147 (45%), Gaps = 11/147 (7%)
Query 7 VTLTREVDG--LKVIVEFSCI----QPAAAAEEEAALPESS-DFTVSVHRSDGSGLLFYC 59
VTLTR+ +G +KV+V + ++E ESS V+V + G L F C
Sbjct 285 VTLTRDYNGEHIKVVVSMPSLVSDENDDDDDDDEGPSNESSIPLVVTVTKKSGLTLEFSC 344
Query 60 STTAEDETARFCIGQVRHFVDSSSKESLASYPGPYFEDLDEAFQQQLDSWLAAVGVDGQL 119
DE A + V+H DS E + GP FEDLDE ++ +L GV
Sbjct 345 -MAFPDEIAIDALS-VKHPGDS--LEDQLANEGPDFEDLDENLKKTFYKFLEIRGVKAST 400
Query 120 CDFLEAFAADKENREYLRWLHKVHAFL 146
+FL + K NREY WL V F+
Sbjct 401 TNFLHEYMTRKVNREYFLWLKNVKEFM 427
> At5g02050
Length=267
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 50/104 (48%), Gaps = 5/104 (4%)
Query 43 FTVSVHRSDGSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESLASYPGPYFEDLDEAF 102
VSV + DG L F S ++ I + S LA Y GP F+DLDE
Sbjct 167 MVVSVQKGDGVCLEFGVSAYPDE----IVIDSLSIKQPQGSDNDLA-YEGPDFDDLDENL 221
Query 103 QQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
Q+ +L G+ FL + A+K++REYL+WL + +F+
Sbjct 222 QKAFHRYLEIRGIKPSFTTFLADYVANKDSREYLQWLKDLKSFV 265
> At5g05990
Length=259
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 71/167 (42%), Gaps = 26/167 (15%)
Query 1 KENDVLVTLTREVDGLKVIVEFSCIQPAAAAEE------------------EAALPESSD 42
K +VTLTRE +G V VE +E + P+ S+
Sbjct 96 KPGGKIVTLTREYEGETVKVEVHMTNLVTGDKEDDEDDEEEAENEEDEDEDKPLKPKQSN 155
Query 43 --FTVSVHRSDGSGLLFYCSTTAEDETARFCIGQV-RHFVDSSSKESLASYPGPYFEDLD 99
V++ + G L F C+ + R I + F D SK+ LA Y GP F LD
Sbjct 156 VPLLVTLSKKTGPSLEFRCTAFPD----RIAIKDMWVTFPDDPSKDELA-YEGPSFRVLD 210
Query 100 EAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
E ++ ++ G+ + +FL + +K++RE+L WL + F+
Sbjct 211 EKLRKAFHRYIEIRGIKPSMINFLHEYMINKDSREHLLWLKSLKNFV 257
> At1g80720
Length=190
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 55/106 (51%), Gaps = 9/106 (8%)
Query 43 FTVSVHRSDGS-GLLFYCSTTAED-ETARFCIGQVRHFVDSSSKESLASYPGPYFEDLDE 100
F V++ ++ + L CS + E ++ CI R +++S +SY GP FE+LD+
Sbjct 89 FIVNISKAGATEALEIMCSAWPDTIEISKLCI---RRGINTSP----SSYGGPEFEELDD 141
Query 101 AFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
Q L +L G+ +L FL + +K EY+RW+ V +++
Sbjct 142 QLQDALYQFLEERGISDELAVFLHRYMKNKGKAEYVRWMESVKSYV 187
> SPBC776.07
Length=269
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 90 YPGPYFEDLDEAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL 146
Y GP F++LD Q S+L +D L F+ +F KE +EY+ WL V FL
Sbjct 212 YLGPSFKELDPELQDLFHSYLEERKIDESLSSFIVSFGLTKELKEYINWLESVRQFL 268
> At4g32610
Length=557
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 33/61 (54%), Gaps = 1/61 (1%)
Query 90 YPGPYFEDLDEAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFL-AN 148
Y GP FE+LDE + +L GV+ L FL+A+ K++R LRW V F+ N
Sbjct 491 YTGPSFEELDEKMRDVFHGFLEERGVNESLFPFLQAWLYVKDHRNLLRWFKSVGTFVHEN 550
Query 149 P 149
P
Sbjct 551 P 551
> Hs4502491
Length=282
Score = 34.7 bits (78), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 44/169 (26%), Positives = 67/169 (39%), Gaps = 24/169 (14%)
Query 3 NDVLVTLTREVDGLKVIVEFSC---IQPAAAAEEEAA-----------LPESSDFTVSVH 48
N L R+V G K+ V F+ I P EEE + L + +F V V
Sbjct 114 NGTEAKLVRKVAGEKITVTFNINNSIPPTFDGEEEPSQGQKVEEQEPELTSTPNFVVEVI 173
Query 49 RSD-GSGLLFYCSTTAEDETAR--------FCIGQVRHFVDSSSKESLASYPGPYFEDLD 99
++D G L EDE + F I +V F + E + + LD
Sbjct 174 KNDDGKKALVLDCHYPEDEVGQEDEAESDIFSIREV-SFQSTGESEWKDTNYTLNTDSLD 232
Query 100 EAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFLAN 148
A L +LA GVD D L + E++EY+ +L + +F+ +
Sbjct 233 WALYDHLMDFLADRGVDNTFADELVELSTALEHQEYITFLEDLKSFVKS 281
> 7299081
Length=979
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 32/73 (43%), Gaps = 1/73 (1%)
Query 37 LPESSDFTVSVHRSDGSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESLASYPGPYFE 96
L S+ F VS DG T D+T +F + Q+ HF D S + + Y +
Sbjct 130 LSLSTSFLVSSGSLDGGNESIKAQTVVADKT-KFRLEQLDHFDDGSMRHMMDLTQQEYIQ 188
Query 97 DLDEAFQQQLDSW 109
++ Q+ + SW
Sbjct 189 RFEQLKQELIQSW 201
> CE18500
Length=1404
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 34/72 (47%), Gaps = 5/72 (6%)
Query 45 VSVHRSDGSGLLFYCSTTAEDET---ARFCIGQVRHFVDSSSKESLASYP--GPYFEDLD 99
+ VH SG+L YCS+ D T FC ++ +++ ++ GP
Sbjct 1198 ILVHFCVKSGVLGYCSSVFADLTPDLTGFCTKLLKTGGSRKKAQNIRNFKNRGPKLGRNS 1257
Query 100 EAFQQQLDSWLA 111
E F+ Q+DSW++
Sbjct 1258 EIFKFQVDSWIS 1269
> CE17940
Length=236
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 109 WLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFLA 147
+L G+D + C L A+A E+ +Y+ L K+ F++
Sbjct 197 YLEERGLDARFCKTLVAYATHYEHSQYVGLLDKIKKFIS 235
> Hs17472020
Length=508
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 8/76 (10%)
Query 24 CIQPAAAAEEEAALPESSDFTVSVHRSDGSGLLFYCSTTAEDETARFCIGQVR------H 77
CI+P A +E + L +++DF + + S L +C + + F + ++R H
Sbjct 374 CIRPCRAQKEASVLLQTNDFNTMLEQC--SSLKDFCPHRSAAHSITFTVRKLRSLDTCGH 431
Query 78 FVDSSSKESLASYPGP 93
+ +KE + S GP
Sbjct 432 RTLTGAKECVRSASGP 447
> SPAC13D6.04c
Length=523
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 32/65 (49%), Gaps = 5/65 (7%)
Query 74 QVRHFVDSSSKESLASYPGPYFEDLDEAFQQQLDSWLAAVGVDGQLCDFLEAFAADKENR 133
+V+HF + ES+ Y + D + F +Q ++ L ++G QL DF+ + D+E
Sbjct 207 EVKHF--AKEFESILRY---LYLDTNAVFTKQYNNALLSIGKKFQLNDFIALYEKDREQL 261
Query 134 EYLRW 138
W
Sbjct 262 HSRDW 266
> AtCh085
Length=2294
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 39/79 (49%), Gaps = 7/79 (8%)
Query 32 EEEAALPESSD-FTVSVHRSD---GSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESL 87
E E E +D FT+S+ D G F + D+ R + +V +F D S K+SL
Sbjct 609 ESEERFQEMADLFTLSITEPDLVYHKGFAFSIDSYGLDQ--RQFLKEVFNFRDESKKKSL 666
Query 88 ASYPGPYFEDLDEAFQQQL 106
P P F + +E+F ++L
Sbjct 667 LVLP-PIFYEENESFYRRL 684
> AtCh065
Length=2294
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 39/79 (49%), Gaps = 7/79 (8%)
Query 32 EEEAALPESSD-FTVSVHRSD---GSGLLFYCSTTAEDETARFCIGQVRHFVDSSSKESL 87
E E E +D FT+S+ D G F + D+ R + +V +F D S K+SL
Sbjct 609 ESEERFQEMADLFTLSITEPDLVYHKGFAFSIDSYGLDQ--RQFLKEVFNFRDESKKKSL 666
Query 88 ASYPGPYFEDLDEAFQQQL 106
P P F + +E+F ++L
Sbjct 667 LVLP-PIFYEENESFYRRL 684
> 7302739
Length=263
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 38/165 (23%), Positives = 62/165 (37%), Gaps = 31/165 (18%)
Query 7 VTLTREVDGLKVIVEFSC-----------IQPAAAAEEEAALPESSDFTVSVHRSDGSGL 55
V LT++ D KV+V F+ I P A + + F V + + + S L
Sbjct 109 VELTKQTDKEKVVVSFNVNHTVDSEEEPEINPNADKPDLGEMRSKPQFEVDIIKGN-STL 167
Query 56 LFYCSTTAEDETARFCIGQVR--HFVDSSSKESLASYPGPYFED--------LDEAFQQQ 105
F CS F G+ + + D S + +A + G + + LD
Sbjct 168 SFTCS---------FLQGEAQEGEYNDVFSIDEMAIFEGEWNDKVYAVAGDVLDGYLYDL 218
Query 106 LDSWLAAVGVDGQLCDFLEAFAADKENREYLRWLHKVHAFLANPK 150
L + L G+ + + L A E+ Y+ L K+ F A K
Sbjct 219 LINLLEEKGISQEFAEKLSDLATAHEHTSYIGLLEKLSKFTAGGK 263
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40