bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1038_orf1
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
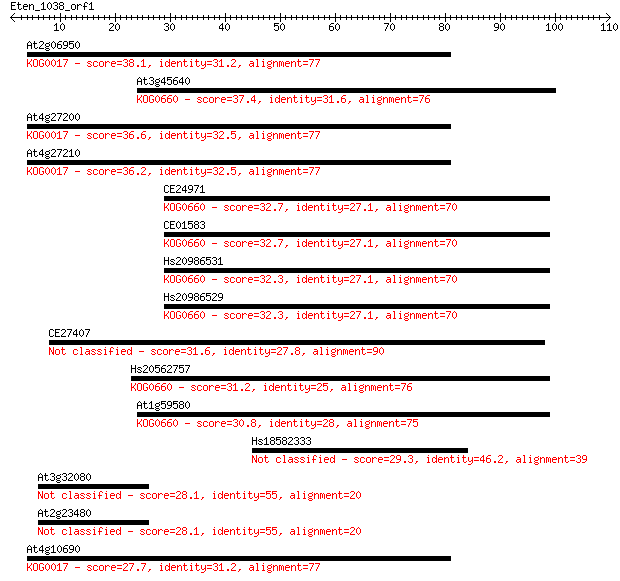
At2g06950 38.1 0.004
At3g45640 37.4 0.007
At4g27200 36.6 0.013
At4g27210 36.2 0.014
CE24971 32.7 0.16
CE01583 32.7 0.16
Hs20986531 32.3 0.22
Hs20986529 32.3 0.22
CE27407 31.6 0.34
Hs20562757 31.2 0.51
At1g59580 30.8 0.59
Hs18582333 29.3 1.8
At3g32080 28.1 3.8
At2g23480 28.1 3.8
At4g10690 27.7 5.4
> At2g06950
Length=1402
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query 4 SVHRPSGTTFHTSGMYLLYLRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLS 63
+H+P+ + FH L Y++ +T+ IS + S S + N C+ TRR +
Sbjct 1240 KMHQPTMSDFHLLKRILRYIKGTITMGISYNQNSPTLLQAYSDSDWGN---CKLTRRSVG 1296
Query 64 GLST--SSSLIAWSSMYRP 80
GL T +++L++WSS P
Sbjct 1297 GLCTFMATNLVSWSSKKHP 1315
> At3g45640
Length=370
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 35/79 (44%), Gaps = 3/79 (3%)
Query 24 RTFLTLFISASLMDLQFHSRL-STSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIH 82
R F ++IS LMD H + S S S +C F + L GL S +P +
Sbjct 110 RQFSDVYISTELMDTDLHQIIRSNQSLSEEHCQYFLYQLLRGLKYIHSANIIHRDLKPSN 169
Query 83 TIM--NCDVNTCSAAVLRP 99
++ NCD+ C + RP
Sbjct 170 LLLNANCDLKICDFGLARP 188
> At4g27200
Length=819
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 4 SVHRPSGTTFHTSGMYLLYLRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLS 63
+H+P+ + FH L Y++ +T+ IS S S S + N C+ TRR +
Sbjct 530 KMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGN---CKQTRRSVG 586
Query 64 GLST--SSSLIAWSSMYRP 80
GL T ++L++WSS P
Sbjct 587 GLCTFMGTNLVSWSSKKHP 605
> At4g27210
Length=1318
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 4 SVHRPSGTTFHTSGMYLLYLRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLS 63
+H+P+ + FH L Y++ +T+ IS S S S + N C+ TRR +
Sbjct 1029 KMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGN---CKQTRRSVG 1085
Query 64 GLST--SSSLIAWSSMYRP 80
GL T ++L++WSS P
Sbjct 1086 GLCTFMGTNLVSWSSKKHP 1104
> CE24971
Length=444
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 29 LFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIHTIMN-- 86
++I LM+ + L T SN + C F + L GL S +P + ++N
Sbjct 172 IYIVQCLMETDLYKLLKTQKLSNDHVCYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTT 231
Query 87 CDVNTCSAAVLR 98
CD+ C + R
Sbjct 232 CDLKICDFGLAR 243
> CE01583
Length=376
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 29 LFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIHTIMN-- 86
++I LM+ + L T SN + C F + L GL S +P + ++N
Sbjct 104 IYIVQCLMETDLYKLLKTQKLSNDHVCYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTT 163
Query 87 CDVNTCSAAVLR 98
CD+ C + R
Sbjct 164 CDLKICDFGLAR 175
> Hs20986531
Length=360
Score = 32.3 bits (72), Expect = 0.22, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 29 LFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIHTIMN-- 86
++I LM+ + L T SN + C F + L GL S +P + ++N
Sbjct 101 VYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTT 160
Query 87 CDVNTCSAAVLR 98
CD+ C + R
Sbjct 161 CDLKICDFGLAR 172
> Hs20986529
Length=360
Score = 32.3 bits (72), Expect = 0.22, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 31/72 (43%), Gaps = 2/72 (2%)
Query 29 LFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIHTIMN-- 86
++I LM+ + L T SN + C F + L GL S +P + ++N
Sbjct 101 VYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTT 160
Query 87 CDVNTCSAAVLR 98
CD+ C + R
Sbjct 161 CDLKICDFGLAR 172
> CE27407
Length=352
Score = 31.6 bits (70), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 43/93 (46%), Gaps = 3/93 (3%)
Query 8 PSGTTFHTSGMYLLYLRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLST 67
P F+ G L Y R+ + + +S + H +T++ +N Y ++ RD S+
Sbjct 150 PPALPFYYRGPELFYYRSKVCITLSRGAGKPRRHCICNTANCNNVYNFQYIARDREKESS 209
Query 68 --SSSLIAWSSMYR-PIHTIMNCDVNTCSAAVL 97
S SL ++ Y P+ T NC+ NT A +
Sbjct 210 LRSRSLTLSATDYTLPLQTCYNCETNTQDATAM 242
> Hs20562757
Length=379
Score = 31.2 bits (69), Expect = 0.51, Method: Composition-based stats.
Identities = 19/78 (24%), Positives = 32/78 (41%), Gaps = 2/78 (2%)
Query 23 LRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIH 82
L ++I LM+ + L + SN + C F + L GL S +P +
Sbjct 112 LEAMRDVYIVQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSN 171
Query 83 TIMN--CDVNTCSAAVLR 98
++N CD+ C + R
Sbjct 172 LLINTTCDLKICDFGLAR 189
> At1g59580
Length=376
Score = 30.8 bits (68), Expect = 0.59, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 35/78 (44%), Gaps = 3/78 (3%)
Query 24 RTFLTLFISASLMDLQFHSRLSTSS-FSNAYCCRFTRRDLSGLSTSSSLIAWSSMYRPIH 82
R+F +++ LMD H + +S SN +C F + L GL S +P +
Sbjct 104 RSFKDVYLVYELMDTDLHQIIKSSQVLSNDHCQYFLFQLLRGLKYIHSANILHRDLKPGN 163
Query 83 TIM--NCDVNTCSAAVLR 98
++ NCD+ C + R
Sbjct 164 LLVNANCDLKICDFGLAR 181
> Hs18582333
Length=127
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 9/48 (18%)
Query 45 STSSFSNAYCCRFTRR---------DLSGLSTSSSLIAWSSMYRPIHT 83
S SS+S+ +C F D LS +S L ++SSM +PIHT
Sbjct 33 SNSSYSHLFCIYFLDHSFSDSKGSTDFPHLSATSFLPSFSSMVKPIHT 80
> At3g32080
Length=370
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 6 HRPSGTTFHTSGMYLLYLRT 25
H PSG T GM++L+LRT
Sbjct 60 HLPSGYTHKLMGMWMLFLRT 79
> At2g23480
Length=705
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 6 HRPSGTTFHTSGMYLLYLRT 25
H PSG T GM++L+LRT
Sbjct 338 HLPSGYTHKLMGMWMLFLRT 357
> At4g10690
Length=1515
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 5/79 (6%)
Query 4 SVHRPSGTTFHTSGMYLLYLRTFLTLFISASLMDLQFHSRLSTSSFSNAYCCRFTRRDLS 63
+H P+ + FH L YL+ +T+ I+ S S L S S+ C+ TRR
Sbjct 1233 KMHAPTMSDFHLLKRILHYLKGTMTMGINLS---SNTDSVLRCYSDSDWAGCKDTRRSTG 1289
Query 64 GLST--SSSLIAWSSMYRP 80
G T ++I+WS+ P
Sbjct 1290 GFCTFLGYNIISWSAKRHP 1308
Lambda K H
0.328 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40