bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1022_orf1
Length=299
Score E
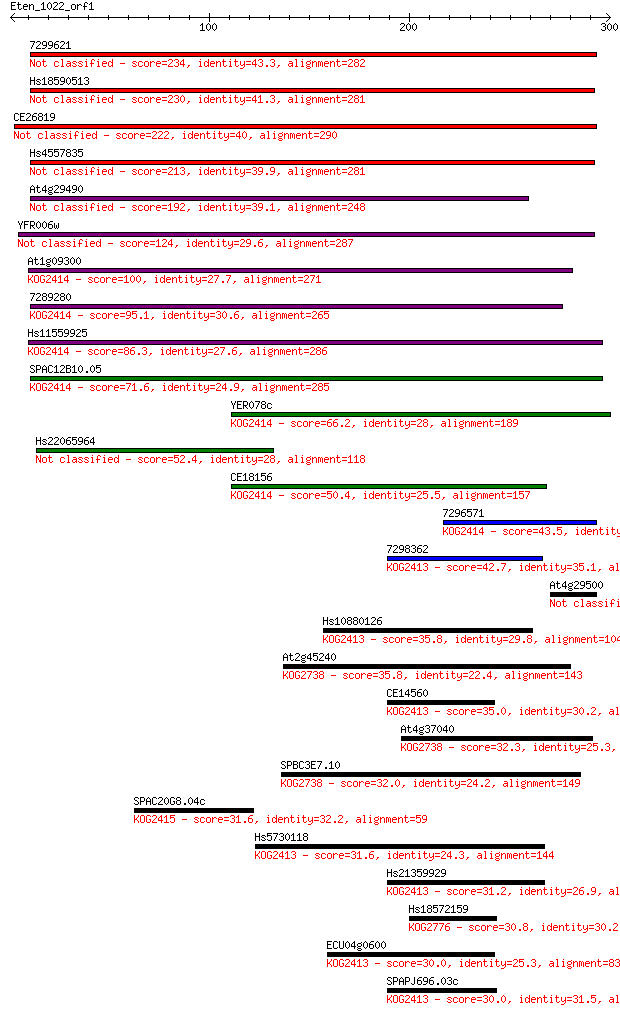
Sequences producing significant alignments: (Bits) Value
7299621 234 1e-61
Hs18590513 230 3e-60
CE26819 222 8e-58
Hs4557835 213 3e-55
At4g29490 192 9e-49
YFR006w 124 2e-28
At1g09300 100 5e-21
7289280 95.1 2e-19
Hs11559925 86.3 8e-17
SPAC12B10.05 71.6 2e-12
YER078c 66.2 8e-11
Hs22065964 52.4 1e-06
CE18156 50.4 4e-06
7296571 43.5 5e-04
7298362 42.7 9e-04
At4g29500 36.2 0.092
Hs10880126 35.8 0.10
At2g45240 35.8 0.12
CE14560 35.0 0.18
At4g37040 32.3 1.3
SPBC3E7.10 32.0 1.5
SPAC20G8.04c 31.6 2.1
Hs5730118 31.6 2.2
Hs21359929 31.2 2.9
Hs18572159 30.8 3.4
ECU04g0600 30.0 6.5
SPAPJ696.03c 30.0 6.5
> 7299621
Length=483
Score = 234 bits (598), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 122/286 (42%), Positives = 171/286 (59%), Gaps = 8/286 (2%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLN----SHELLLFIPKIPADALRFMGPSKDADFYRSR 66
FRQE++FQ+L GV EP Y ++ + + +LF+P+ P + +MG +++
Sbjct 64 FRQESYFQYLFGVKEPGCYGILTIDVKTGAQKSVLFVPRFPDEYGTWMGELLGLQEFKAM 123
Query 67 YAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSAL 126
Y V + V+ +S L+ + + L G NSDSG + PP + DC+ L
Sbjct 124 YEVDEVFYVDEMS---VYLEGASPKLILTLSGTNSDSGLTLQPPDFAGK-EKYVTDCNLL 179
Query 127 YDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGA 186
Y IL ECR+ K+P E E L SS AH V R +R G +E + E++F + VGG
Sbjct 180 YPILSECRVIKSPEEIEVLRYVAKVSSDAHIKVMRFMRPGRMEFEGESLFLHHAYSVGGC 239
Query 187 RHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNG 246
RH +Y CIC +G N+SILHYGHAG PN ++D DL LFDMG +Y GYA DIT T+P+NG
Sbjct 240 RHASYTCICGSGTNSSILHYGHAGAPNSKPVQDGDLCLFDMGANYCGYAADITCTFPANG 299
Query 247 IFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
FT+ QK IY+AVLDA+ +V E R GV W ++H+LA V+L+ LK
Sbjct 300 KFTDDQKFIYNAVLDARNAVTESARDGVSWVDMHKLAGRVLLQRLK 345
> Hs18590513
Length=493
Score = 230 bits (586), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 116/281 (41%), Positives = 178/281 (63%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE+FF GV EP Y +++ + LF+P++PA +MG + ++ +YAV
Sbjct 65 FRQESFFHWAFGVTEPGCYGVIDVDTGKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVD 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
D V+ I+ V L + + L+GVN+DSG V + + S F V+ + L+ +
Sbjct 125 DVQYVDEIASV---LTSQKPSVLLTLRGVNTDSGS-VCREASFDGISKFEVNNTILHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VECR+ KT +E E L SS+AH V + ++ GM E + E++F+ Y GG RH +
Sbjct 181 VECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESLFEHYCYSRGGMRHSS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CIC +GEN+++LHYGHAG PND +I++ D+ LFDMGG+Y +A+DIT ++P+NG FT
Sbjct 241 YTCICGSGENSAVLHYGHAGAPNDRTIQNGDMCLFDMGGEYYCFASDITCSFPANGKFTA 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
QKA+Y+AVL + ++V+ M+PGV W ++HRLA+ + L+ L
Sbjct 301 DQKAVYEAVLRSSRAVMGAMKPGVWWPDMHRLADRIHLEEL 341
> CE26819
Length=498
Score = 222 bits (565), Expect = 8e-58, Method: Compositional matrix adjust.
Identities = 116/291 (39%), Positives = 173/291 (59%), Gaps = 2/291 (0%)
Query 3 SADCNKAAFRQEAFFQHLVGVNEPDIYAAF-VLNSHELLLFIPKIPADALRFMGPSKDAD 61
+ D FRQE++F GVNE + Y A V + + LF P++ + G +
Sbjct 50 NTDAADLPFRQESYFFWTFGVNESEFYGAIDVRSGGKTTLFAPRLDPSYAIWDGKINNEQ 109
Query 62 FYRSRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTV 121
F++ +YAV + + + + + +L+ + V++L+ N+DSG +A PK S F +
Sbjct 110 FFKEKYAVDEVVFNDKTTTIAEKLKELSAKHVYLLRAENTDSGDVLAEPKFAGS-GDFQL 168
Query 122 DCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVG 181
D LY + E R+ KT E + A +S+AH +++R G+ E Q E++F+
Sbjct 169 DTELLYKEMAELRVVKTEKEIGVMRYASKIASEAHRAAMKHMRPGLYEYQLESLFRHTSY 228
Query 182 FVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFT 241
+ GG RH+AY CI G N S+LHYGHA PND IKD D+ LFDMG +YN YA+DIT +
Sbjct 229 YHGGCRHLAYTCIAATGCNGSVLHYGHANAPNDKFIKDGDMCLFDMGPEYNCYASDITTS 288
Query 242 YPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLK 292
+PSNG FTE QK +Y+AVL A +V++ +PGV WT++H L+E VIL+HLK
Sbjct 289 FPSNGKFTEKQKIVYNAVLAANLAVLKAAKPGVRWTDMHILSEKVILEHLK 339
> Hs4557835
Length=493
Score = 213 bits (543), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 112/281 (39%), Positives = 171/281 (60%), Gaps = 4/281 (1%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
F QE+FF GV EP Y +++ + LF+P++PA +MG + ++ +YAV
Sbjct 65 FLQESFFHWAFGVTEPGCYGVIDVDTGKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVD 124
Query 71 DAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDIL 130
D V+ I+ V L + + L+GVN+DSG V + + S F V+ + L+ +
Sbjct 125 DVQYVDEIASV---LTSQKPSVLLTLRGVNTDSGS-VCREASFDGISKFEVNNTILHPEI 180
Query 131 VECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHVA 190
VE R+ KT +E E L SS+AH V + ++ GM E E++F+ Y GG RH +
Sbjct 181 VESRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYGLESLFEHYCYSRGGMRHSS 240
Query 191 YDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTE 250
Y CIC +GEN+++LHYGHAG PND +I++ D+ LFDMGG+Y A+DIT ++P NG FT
Sbjct 241 YTCICGSGENSAVLHYGHAGAPNDRTIQNGDMCLFDMGGEYYSVASDITCSFPRNGKFTA 300
Query 251 TQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
QKA+Y+AVL + ++V+ M+PG W ++ RLA+ + L+ L
Sbjct 301 DQKAVYEAVLLSSRAVMGAMKPGDWWPDIDRLADRIHLEEL 341
> At4g29490
Length=333
Score = 192 bits (487), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 97/249 (38%), Positives = 143/249 (57%), Gaps = 2/249 (0%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVT 70
FRQE++F +L GV EPD Y A + S + +LFIP++P D ++G K ++ Y V
Sbjct 59 FRQESYFAYLFGVREPDFYGAIDIGSGKSILFIPRLPDDYAVWLGEIKPLSHFKETYMVD 118
Query 71 DAIAVEAISDV-EAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDI 129
V+ I V + + G +++L G+N+DS + P + F D + L+ I
Sbjct 119 MVFYVDEIIQVFNEQFKGSGKPLLYLLHGLNTDSSN-FSKPASFEGIDKFETDLTTLHPI 177
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVGGARHV 189
L ECR+ K+ LE + + A SS+AH V R + GM E Q E++F + GG RH
Sbjct 178 LAECRVIKSSLELQLIQFANDISSEAHIEVMRKVTPGMKEYQMESMFLHHSYMYGGCRHC 237
Query 190 AYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFT 249
+Y CIC G+N+++LHYGHA PND + +D DL L DMG +Y+ Y +DIT ++P NG FT
Sbjct 238 SYTCICATGDNSAVLHYGHAAAPNDRTFEDGDLALLDMGAEYHFYGSDITCSFPVNGKFT 297
Query 250 ETQKAIYDA 258
Q IY+
Sbjct 298 SDQSLIYNV 306
> YFR006w
Length=535
Score = 124 bits (312), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 85/287 (29%), Positives = 134/287 (46%), Gaps = 18/287 (6%)
Query 5 DCNKAAFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYR 64
D NK FRQ +F HL GV+ P F ++ +L LF+P I + + + G D
Sbjct 113 DTNKD-FRQNRYFYHLSGVDIPASAILFNCSTDKLTLFLPNIDEEDVIWSGMPLSLDEAM 171
Query 65 SRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCS 124
+ + +A+ + + ELQ + T + + + R + P D +
Sbjct 172 RVFDIDEALYISDLGKKFKELQDFAIFTTDLDNVHDENIARSLIPS-----------DPN 220
Query 125 ALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGFVG 184
Y + E R K E E + AC S ++H V + + E Q +A F+ + G
Sbjct 221 FFY-AMDETRAIKDWYEIESIRKACQISDKSHLAVMSALPIELNELQIQAEFEYHATRQG 279
Query 185 GARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPS 244
G R + YD ICC+G LHY N IK +L D G ++ Y +DIT +P+
Sbjct 280 G-RSLGYDPICCSGPACGTLHYVK----NSEDIKGKHSILIDAGAEWRQYTSDITRCFPT 334
Query 245 NGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHL 291
+G FT + +Y+ VLD Q +ER++PG +W +LH L V++KH
Sbjct 335 SGKFTAEHREVYETVLDMQNQAMERIKPGAKWDDLHALTHKVLIKHF 381
> At1g09300
Length=451
Score = 100 bits (248), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 75/277 (27%), Positives = 127/277 (45%), Gaps = 23/277 (8%)
Query 10 AFRQEAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAV 69
FRQ+A + +L G +P A + + L +F+P+ + + G D +
Sbjct 49 TFRQDADYLYLTGCQQPG-GVAVLSDERGLCMFMPESTPKDIAWEGEVAGVDAASEVFKA 107
Query 70 TDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDI 129
A + + ++ +++ R + H V S S R NS S V L +
Sbjct 108 DQAYPISKLPEILSDMIRHSSKVFH---NVQSASQRYTNLDDFQNSASLGKV--KTLSSL 162
Query 130 LVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQA---EAIFKAYVGF---V 183
E RL K+P E + + + + Q + M+ + E I A V + V
Sbjct 163 THELRLIKSPAELKLMRESASIACQG-------LLKTMLHSKGFPDEGILSAQVEYECRV 215
Query 184 GGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYP 243
GA+ +A++ + G NAS++HY ND IKD DL+L DMG + +GY +D+T T+P
Sbjct 216 RGAQRMAFNPVVGGGSNASVIHYSR----NDQRIKDGDLVLMDMGCELHGYVSDLTRTWP 271
Query 244 SNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELH 280
G F+ Q+ +YD +L K I++ +PG +L+
Sbjct 272 PCGKFSSVQEELYDLILQTNKECIKQCKPGTTIRQLN 308
> 7289280
Length=545
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 81/273 (29%), Positives = 123/273 (45%), Gaps = 20/273 (7%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVL----NSHELLLFIPKIPADALRFMGPSKDADFYRSR 66
FRQ + F +L G EPD + N L PK P L + GP +
Sbjct 143 FRQNSDFYYLTGCLEPDAVLLLTIDEAQNVQSELFMRPKDPHAEL-WDGPRTGPELAVPL 201
Query 67 YAVTDAIAVEAISDVEAELQRR-GVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSA 125
+ VT+A +S +EA L +R G H+ S + P L +
Sbjct 202 FGVTEA---HPLSQLEAVLAKRAGALKPHIWFDQKS-TDLPSLAENMLRLSGNQQRPLLP 257
Query 126 LYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKA--YVGFV 183
Y L RL K+ E + + C +S++ V R G E +F A Y +
Sbjct 258 AYTFLEAMRLLKSRDEMQLMRRTCDIASRSFNEVMAETRPGQSE---HHLFAAIDYKCRM 314
Query 184 GGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYP 243
A ++AY + AG+NA+++HY N + DL+L D G +Y GY +DIT T+P
Sbjct 315 RNASYLAYPPVVAAGKNATVIHY----VANSQLLGQQDLVLMDAGCEYGGYTSDITRTWP 370
Query 244 SNGIFTETQKAIYDAVLDAQKSVIER-MRPGVE 275
++G+FTE Q+ +YD + Q+ +I M+PG E
Sbjct 371 ASGVFTEPQRTLYDMLHQLQEEIIGNVMKPGGE 403
> Hs11559925
Length=507
Score = 86.3 bits (212), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 79/308 (25%), Positives = 134/308 (43%), Gaps = 34/308 (11%)
Query 10 AFRQEAFFQHLVGVNEPDIYAAFV------LNSHELLLFIPKIPADALRFMGPSKDADFY 63
F Q+ F +L G EPD L SH+ +LF+P+ + GP D
Sbjct 114 TFHQDNNFLYLCGFQEPDSILVLQSLPGKQLPSHKAILFVPRRDPSRELWDGPRSGTDGA 173
Query 64 RSRYAVTDAIAVEAISDVEAELQRRG-------VRTVHVLKGVNSDSGRPVAPPKALNSF 116
+ V +A +E + +++ +R H ++SD +P+ KA +
Sbjct 174 IALTGVDEAYTLEEFQHLLPKMKAETNMVWYDWMRPSHA--QLHSDYMQPLTEAKAKSKN 231
Query 117 SSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIF 176
+ ++ RL K+P E E + A +SQA ++ + E A F
Sbjct 232 K-----VRGVQQLIQRLRLIKSPAEIERMQIAGKLTSQAFIETMFTSKAPVEEAFLYAKF 286
Query 177 KAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYAT 236
+ + GA +AY + G ++ LHY N+ IKD +++L D G + + Y +
Sbjct 287 E-FECRARGADILAYPPVVAGGNRSNTLHYVK----NNQLIKDGEMVLLDGGCESSCYVS 341
Query 237 DITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELH---------RLAELVI 287
DIT T+P NG FT Q +Y+AVL+ Q+ + PG ++ +L +L I
Sbjct 342 DITRTWPVNGRFTAPQAELYEAVLEIQRDCLALCFPGTSLENIYSMMLTLIGQKLKDLGI 401
Query 288 LKHLKVRN 295
+K++K N
Sbjct 402 MKNIKENN 409
> SPAC12B10.05
Length=486
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 71/293 (24%), Positives = 124/293 (42%), Gaps = 16/293 (5%)
Query 11 FRQEAFFQHLVGVNEPDIYAAFVLN----SHELLLFIPKIPADALRFMGPSKDADFYRSR 66
+ Q+ F +L G EP+ N S++ L++P ++ G + +
Sbjct 102 YHQDPNFYYLTGCLEPNAVLLMFKNGASGSYDCSLYLPSKNPYIEKWEGLRTGSTLGKKL 161
Query 67 YAVTDAIAVEAISDVEAELQRRGVRTVHVLK-GVNSDSGRPVAPPKALNSFSSF--TVDC 123
+ + + S V L ++ R + + G S AP ++ + T
Sbjct 162 FQIENVYDSSLASSVINALGKKSNRIFYNYQTGYLSKMPAASAPEFIQDTLTKLFRTSTQ 221
Query 124 SALYDILVECRLRKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYVGF- 182
++ ++L R K+ E E + A SS V R I E +AE + F
Sbjct 222 RSVDELLHPLRSIKSTAELECMKEAANISSN----VYREIMRKRFEKEAEMSAEFNYRFC 277
Query 183 VGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTY 242
+GG AY + G+N +HY N+ + ++++L D GG++ GY TDI+ T+
Sbjct 278 IGGCDRSAYVPVVAGGKNGLTIHY----TINNDIFRPDEMVLVDAGGEFGGYVTDISRTW 333
Query 243 PSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVILKHLKVRN 295
P NG F+ Q+ +Y AVL+ QK I+ W+ E V L H +++
Sbjct 334 PINGKFSTVQRDLYQAVLNVQKKCIKYCCTSNGWSLADIHFESVKLMHEELKQ 386
> YER078c
Length=511
Score = 66.2 bits (160), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 53/197 (26%), Positives = 92/197 (46%), Gaps = 20/197 (10%)
Query 111 KALNSFSSFTVDCSALYDILVECRLRKTPLEAEYLAAACLCSSQA--HCFVARNIRSGMV 168
++LNS ++ T+ + + E R K+P E + A S ++ F R +
Sbjct 221 RSLNSIANKTI--KPISKRIAEFRKIKSPQELRIMRRAGQISGRSFNQAFAKRFRNERTL 278
Query 169 EGQAEAIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMG 228
+ F Y GG AY + G N+ +HY ND + D++++L D
Sbjct 279 DS-----FLHYKFISGGCDKDAYIPVVATGSNSLCIHYTR----NDDVMFDDEMVLVDAA 329
Query 229 GDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMRPGVEWTELHRLAELVI- 287
G GY DI+ T+P++G FT+ Q+ +Y+AVL+ Q+ I+ + ++ LH + E I
Sbjct 330 GSLGGYCADISRTWPNSGKFTDAQRDLYEAVLNVQRDCIKLCKASNNYS-LHDIHEKSIT 388
Query 288 -----LKHLKVRNDTLW 299
LK+L + + W
Sbjct 389 LMKQELKNLGIDKVSGW 405
> Hs22065964
Length=299
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 57/118 (48%), Gaps = 4/118 (3%)
Query 14 EAFFQHLVGVNEPDIYAAFVLNSHELLLFIPKIPADALRFMGPSKDADFYRSRYAVTDAI 73
E+FF GV EP Y ++ + LF+P++PA +MG + ++ +YAV D
Sbjct 186 ESFFHCAFGVTEPGCYGFIDADTGKSALFVPRLPASHATWMGKIHSKEHFKEKYAVDD-- 243
Query 74 AVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTVDCSALYDILV 131
V+ ++ L + + L+GVN+DS V + S F V+ + L+ +V
Sbjct 244 -VQDADEIAIVLTSQKPSVLLTLRGVNTDSSS-VCREASFEGISKFKVNNTILHPEIV 299
> CE18156
Length=383
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 40/160 (25%), Positives = 69/160 (43%), Gaps = 8/160 (5%)
Query 111 KALNSFSSFTVDCSALYDILVECRLR---KTPLEAEYLAAACLCSSQAHCFVARNIRSGM 167
K + F F S L E R K+P E + C +Q + R
Sbjct 93 KKKSDFRGFQSKNSNFVQFLNEIERRRVIKSPSEMSSMRDVCNVGAQTMSSMISGSRDLH 152
Query 168 VEGQAEAIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDM 227
E + + + G G+ AY + G A+ +HY A ND + + +L D
Sbjct 153 NENAICGLLE-FEGRRRGSEMQAYPPVIAGGVRANTIHYLDAN--ND--LNPRECVLVDA 207
Query 228 GGDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVI 267
G D NGY +D+T +P +G +++ Q ++Y+A+L + ++
Sbjct 208 GCDLNGYVSDVTRCFPISGFWSDAQLSLYEALLYVHEELL 247
> 7296571
Length=767
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 6/82 (7%)
Query 217 IKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVLDAQKSVIERMR-PGVE 275
+ N L D G Y GY + +PS+G FT QK IY A+LD ++ + ++ G E
Sbjct 553 LDGNGLWQMDAGCQYGGYEGGLARCWPSSGRFTPPQKMIYGALLDMRRDLCSLIQYAGCE 612
Query 276 WT-----ELHRLAELVILKHLK 292
ELH +++ +HL+
Sbjct 613 GAVRTPLELHAAYLILLARHLR 634
> 7298362
Length=613
Score = 42.7 bits (99), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIF 248
+++ I +G N S++HY H + + I D ++ L D G Y TD+T T G
Sbjct 372 LSFTTISASGPNGSVIHY-HPKKETNRKINDKEIYLCDSGAQYLDGTTDVTRTL-HFGEP 429
Query 249 TETQKAIYDAVLDAQKS 265
TE QK Y VL Q S
Sbjct 430 TEFQKEAYTRVLKGQLS 446
> At4g29500
Length=169
Score = 36.2 bits (82), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 270 MRPGVEWTELHRLAELVILKHLK 292
M+PGV W ++H+LAE +IL+ LK
Sbjct 1 MKPGVNWVDMHKLAEKIILESLK 23
> Hs10880126
Length=674
Score = 35.8 bits (81), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 49/108 (45%), Gaps = 9/108 (8%)
Query 157 CFVARNIRSGMVE--GQAEAI--FKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRP 212
++ +N+ G V+ AE + F+ F G +++ I +G NA++ HY
Sbjct 381 VWLEKNVPKGTVDEFSGAEIVDKFRGEEQFSSGP---SFETISASGLNAALAHYSPTKEL 437
Query 213 NDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAIYDAVL 260
N + +++ L D GG Y TDIT T G + QK Y VL
Sbjct 438 NR-KLSSDEMYLLDSGGQYWDGTTDITRTV-HWGTPSAFQKEAYTRVL 483
> At2g45240
Length=398
Score = 35.8 bits (81), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 32/147 (21%), Positives = 59/147 (40%), Gaps = 9/147 (6%)
Query 137 KTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFK----AYVGFVGGARHVAYD 192
KTP + + + C + + AR I G+ + + + A G+ + +
Sbjct 142 KTPEQIQRMRETCKIAREVLDAAARVIHPGVTTDEIDRVVHEATIAAGGYPSPLNYYFFP 201
Query 193 CICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQ 252
CC N I H G P+ ++D D++ D+ Y G D+ TY G E
Sbjct 202 KSCCTSVNEVICH----GIPDARKLEDGDIVNVDVTVCYKGCHGDLNETY-FVGNVDEAS 256
Query 253 KAIYDAVLDAQKSVIERMRPGVEWTEL 279
+ + + + I ++PGV + E+
Sbjct 257 RQLVKCTYECLEKAIAIVKPGVRFREI 283
> CE14560
Length=616
Score = 35.0 bits (79), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFT 241
+++D I G++A++ HY G + N + L D G Y TD+T T
Sbjct 376 LSFDTISAVGDHAALPHYKPLGESGNRKAAANQVFLLDSGAHYGDGTTDVTRT 428
> At4g37040
Length=305
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 42/95 (44%), Gaps = 5/95 (5%)
Query 196 CAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTETQKAI 255
C N I H G P+ ++D D++ D+ NGY D + T+ G E K +
Sbjct 126 CTSVNECICH----GIPDSRPLEDGDIINIDVTVYLNGYHGDTSATF-FCGNVDEKAKKL 180
Query 256 YDAVLDAQKSVIERMRPGVEWTELHRLAELVILKH 290
+ ++ I PGVE+ ++ ++ + KH
Sbjct 181 VEVTKESLDKAISICGPGVEYKKIGKVIHDLADKH 215
> SPBC3E7.10
Length=379
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 36/159 (22%), Positives = 60/159 (37%), Gaps = 18/159 (11%)
Query 136 RKTPLEAEYLAAACLCSSQAHCFVARNIRSGMVEGQAEAIFKAYV----GFVGGARHVAY 191
R TP E E + C + A +R G + ++I F + A+
Sbjct 119 RLTPKEQEGMRKVCRLGREVLDAAAAAVRPGTTTDELDSIVHNACIERDCFPSTLNYYAF 178
Query 192 DCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIFTET 251
C N I H G P+ ++D D++ D+ +NG+ D+ TY +
Sbjct 179 PKSVCTSVNEIICH----GIPDQRPLEDGDIVNIDVSLYHNGFHGDLNETY----YVGDK 230
Query 252 QKAIYDAVLDAQKS------VIERMRPGVEWTELHRLAE 284
KA D V + + I ++PGV + E + E
Sbjct 231 AKANPDLVCLVENTRIALDKAIAAVKPGVLFQEFGNIIE 269
> SPAC20G8.04c
Length=632
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 9/59 (15%)
Query 63 YRSRYAVTDAIAVEAISDVEAELQRRGVRTVHVLKGVNSDSGRPVAPPKALNSFSSFTV 121
Y SR A+ ++ + ++ V ++ RRG R++HVL P+A P L SS T+
Sbjct 3 YLSRSALARSVGAKHLTGVLRKIGRRGGRSMHVL---------PLASPSTLLKISSQTL 52
> Hs5730118
Length=623
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 35/153 (22%), Positives = 61/153 (39%), Gaps = 15/153 (9%)
Query 123 CSALYDILVECRLRKTPLEAEYLAAACLCSSQAHC----FVARNIRSGMVE-----GQAE 173
C Y + + K E+E + A + + A C ++ + + G V +AE
Sbjct 308 CCMPYTPICIAKAVKNSAESEGMRPAHIKDAVALCELFNWLEKEVPKGGVTEISAADKAE 367
Query 174 AIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNG 233
+ FV +++ I G N +I+HY N ++ +++ L D G Y
Sbjct 368 EFRRQQADFVD----LSFPTISSTGPNGAIIHYAPVPETNR-TLSLDEVYLIDSGAQYKD 422
Query 234 YATDITFTYPSNGIFTETQKAIYDAVLDAQKSV 266
TD+T T G T +K + VL +V
Sbjct 423 GTTDVTRTM-HFGTPTAYEKECFTYVLKGHIAV 454
> Hs21359929
Length=623
Score = 31.2 bits (69), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 35/78 (44%), Gaps = 2/78 (2%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTYPSNGIF 248
+++ I G N +I+HY N ++ +++ L D G Y TD+T T G
Sbjct 379 LSFPTISSTGPNGAIIHYAPVPETNR-TLSLDEVYLIDSGAQYKDGTTDVTRTM-HFGTP 436
Query 249 TETQKAIYDAVLDAQKSV 266
T +K + VL +V
Sbjct 437 TAYEKECFTYVLKGHIAV 454
> Hs18572159
Length=910
Score = 30.8 bits (68), Expect = 3.4, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 200 NASILHYGHAGRPNDGSIKDNDLLLFDMGGDYNGYATDITFTY 242
N + H+ R D +K+ DL+ D+G NG+ T++ T+
Sbjct 491 NNCVCHFSPLKRDQDYILKEGDLVKIDLGVHVNGFITNVARTF 533
> ECU04g0600
Length=586
Score = 30.0 bits (66), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 39/83 (46%), Gaps = 8/83 (9%)
Query 159 VARNIRSGMVEGQAEAIFKAYVGFVGGARHVAYDCICCAGENASILHYGHAGRPNDGSIK 218
+ + I V+ + + I K + G+V +++ I G N +I+H+ + D +
Sbjct 334 LEKGISEKDVKEKLDEIKKRFSGYV----QPSFESIVGGGPNGAIVHH----KAGDRIMS 385
Query 219 DNDLLLFDMGGDYNGYATDITFT 241
++L+L D G Y TD T T
Sbjct 386 RDELILIDSGSQYMFGTTDTTRT 408
> SPAPJ696.03c
Length=242
Score = 30.0 bits (66), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 27/56 (48%), Gaps = 5/56 (8%)
Query 189 VAYDCICCAGENASILHYGHAGRPNDGS--IKDNDLLLFDMGGDYNGYATDITFTY 242
++++ I G N +++HY P GS I + L D G Y TD+T T+
Sbjct 5 LSFETISSTGPNGAVIHYS---PPATGSAIIDPTKIYLCDSGAQYKDGTTDVTRTW 57
Lambda K H
0.322 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6743279066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40