bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1018_orf2
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
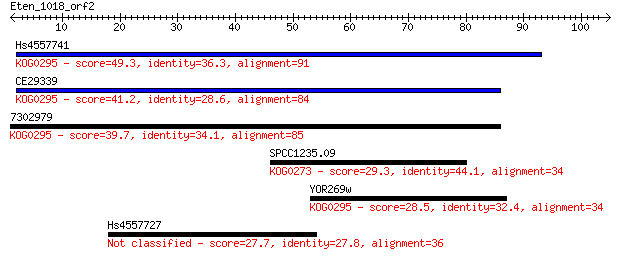
Hs4557741 49.3 2e-06
CE29339 41.2 4e-04
7302979 39.7 0.001
SPCC1235.09 29.3 2.0
YOR269w 28.5 3.3
Hs4557727 27.7 4.7
> Hs4557741
Length=410
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 33/95 (34%), Positives = 51/95 (53%), Gaps = 16/95 (16%)
Query 2 MALRARQEQQLHLAIALYLESK-FPEAYRVFEREAHVDFSAATALTHGSEKFDSE---TL 57
M L RQ +L+ AIA YL S + EAY VF++EA +D + E+ D + L
Sbjct 1 MVLSQRQRDELNRAIADYLRSNGYEEAYSVFKKEAELDVN---------EELDKKYAGLL 51
Query 58 PKRWGATARLQARVTVLQNQLQQQGQQLKLFTSAA 92
K+W + RLQ +V L+++L + ++ FTS
Sbjct 52 EKKWTSVIRLQKKVMELESKLNEAKEE---FTSGG 83
> CE29339
Length=404
Score = 41.2 bits (95), Expect = 4e-04, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 49/85 (57%), Gaps = 9/85 (10%)
Query 2 MALRARQEQQLHLAIALYLESK-FPEAYRVFEREAHVDFSAATALTHGSEKFDSETLPKR 60
M+L RQ+++++ AIA Y+++ + E++ VF +E ++L+ K L K+
Sbjct 1 MSLSERQKEEINRAIAEYMQNNGYSESFSVFLKE--------SSLSENDIKPLGGILEKK 52
Query 61 WGATARLQARVTVLQNQLQQQGQQL 85
W RLQ +V L+++LQ+ +++
Sbjct 53 WTTVLRLQRKVNDLESKLQESQREI 77
> 7302979
Length=411
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 46/86 (53%), Gaps = 9/86 (10%)
Query 1 KMALRARQEQQLHLAIALYLESK-FPEAYRVFEREAHVDFSAATALTHGSEKFDSETLPK 59
KM L RQ ++L+ AIA YL S + ++ F +EA D S T +KF L K
Sbjct 2 KMVLSQRQREELNQAIADYLGSNGYADSLETFRKEA--DLS-----TEVEKKFGG-LLEK 53
Query 60 RWGATARLQARVTVLQNQLQQQGQQL 85
+W + RLQ +V L+ +L + +++
Sbjct 54 KWTSVIRLQKKVMELEAKLTEAEKEV 79
> SPCC1235.09
Length=564
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 4/38 (10%)
Query 46 THGSEKFDSET----LPKRWGATARLQARVTVLQNQLQ 79
+H F+ ET L K+WG+T ++ A V +LQ LQ
Sbjct 20 SHTKFAFERETGIQNLDKQWGSTCQVGALVEILQKGLQ 57
> YOR269w
Length=494
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 53 DSETLPKRWGATARLQARVTVLQNQLQQQGQQLK 86
D LPK+W + RLQ ++ L+ + Q+K
Sbjct 81 DELLLPKKWNSIVRLQKKIIELEQNTETLVSQIK 114
> Hs4557727
Length=475
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 10/36 (27%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 18 LYLESKFPEAYRVFEREAHVDFSAATALTHGSEKFD 53
+YL+++ Y+VF + + FS + TH ++ F+
Sbjct 328 MYLKTRSQMPYKVFHYQVKIHFSGTESETHTNQAFE 363
Lambda K H
0.318 0.127 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40