bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1011_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
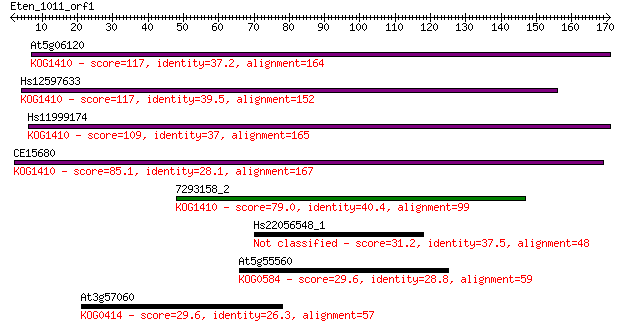
At5g06120 117 7e-27
Hs12597633 117 1e-26
Hs11999174 109 2e-24
CE15680 85.1 6e-17
7293158_2 79.0 5e-15
Hs22056548_1 31.2 0.98
At5g55560 29.6 2.9
At3g57060 29.6 3.0
> At5g06120
Length=1059
Score = 117 bits (294), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 61/165 (36%), Positives = 101/165 (61%), Gaps = 4/165 (2%)
Query 7 CLSCLVLISAFRRSFFETTAIRNKYMGELILGTFQIIKTRTGLSDDMCYHEFCRLIGKVN 66
L CLV +++ RRS F A R+ ++ L+ GT +I++T GL+D YH FCRL+G+
Sbjct 265 ALECLVRLASVRRSLFTNDATRSNFLAHLMTGTKEILQTGKGLADHDNYHVFCRLLGRFR 324
Query 67 TSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHYLLGVWAHMISPLLFYKQRV 126
++ LSEL E + E+ Q + +FT++SL+SWQ +S +YLLG+W+ +++ + + K
Sbjct 325 LNYQLSELVKMEGYGEWIQLVAEFTLKSLQSWQWASSSVYYLLGMWSRLVASVPYLKGDS 384
Query 127 PKDLELYIERITAAFIMSRMCVAEAIAEDSDSCDWESPLNN-EVL 170
P L+ ++ +IT FI+SR +A D + + PL+ EVL
Sbjct 385 PSLLDEFVPKITEGFIISRFNSVQASVPDDPT---DHPLDKVEVL 426
> Hs12597633
Length=1088
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 60/152 (39%), Positives = 89/152 (58%), Gaps = 1/152 (0%)
Query 4 ARLCLSCLVLISAFRRSFFETTAIRNKYMGELILGTFQIIKTRTGLSDDMCYHEFCRLIG 63
++L LSCLV ++ RRS F + R KY+G LI G +I++ GLSD YHEFCR +
Sbjct 276 SQLALSCLVQFASTRRSLFNSPE-RAKYLGNLIKGVKRILENPQGLSDPGNYHEFCRFLA 334
Query 64 KVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHYLLGVWAHMISPLLFYK 123
++ T++ L EL + + E + + +FT+ SL+ W+ PNS HYLL +W M++ + F K
Sbjct 335 RLKTNYQLGELVMVKEYPEVIRLIANFTITSLQHWEFAPNSVHYLLTLWQRMVASVPFVK 394
Query 124 QRVPKDLELYIERITAAFIMSRMCVAEAIAED 155
P L+ Y IT AFI SR+ + D
Sbjct 395 STEPHLLDTYAPEITKAFITSRLDSVAIVVRD 426
> Hs11999174
Length=1087
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 61/165 (36%), Positives = 91/165 (55%), Gaps = 5/165 (3%)
Query 6 LCLSCLVLISAFRRSFFETTAIRNKYMGELILGTFQIIKTRTGLSDDMCYHEFCRLIGKV 65
L LSCLV I++ RRS F A R K++ L+ G +I++ LSD YHEFCRL+ ++
Sbjct 279 LVLSCLVQIASVRRSLF-NNAERAKFLSHLVDGVKRILENPQSLSDPNNYHEFCRLLARL 337
Query 66 NTSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHYLLGVWAHMISPLLFYKQR 125
+++ L EL E + E + + +FT+ SL+ W+ PNS HYLL +W + + + + K
Sbjct 338 KSNYQLGELVKVENYPEVIRLIANFTVTSLQHWEFAPNSVHYLLSLWQRLAASVPYVKAT 397
Query 126 VPKDLELYIERITAAFIMSRMCVAEAIAEDSDSCDWESPLNNEVL 170
P LE Y +T A+I SR+ I D E PL + L
Sbjct 398 EPHMLETYTPEVTKAYITSRLESVHIILRDG----LEDPLEDTGL 438
> CE15680
Length=1078
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 47/167 (28%), Positives = 88/167 (52%), Gaps = 5/167 (2%)
Query 2 ECARLCLSCLVLISAFRRSFFETTAIRNKYMGELILGTFQIIKTRTGLSDDMCYHEFCRL 61
E + ++ + +++ RR+ F T R Y+ +L+ G +I LSD +HEFCRL
Sbjct 278 ESSEKVMTIIAQLASIRRTLFNGTE-RQAYVQKLVEGVVSVIMNPGKLSDQAAFHEFCRL 336
Query 62 IGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHYLLGVWAHMISPLLF 121
I ++ T++ L EL A + + L +FT++SL+ + NS ++L+ W M++ + +
Sbjct 337 IARLKTNYQLCELIAVPCYSHMLRLLAEFTVQSLRMMEFSANSTYFLMTFWQRMVTSVPY 396
Query 122 YKQRVPKDLELYIERITAAFIMSRMCVAEAIAEDSDSCDWESPLNNE 168
+ L +Y I AF+ SR+ E+I + E+PL+++
Sbjct 397 VRNNDEHLLNVYCPEIMTAFVESRLQHVESIVREG----AENPLDDQ 439
> 7293158_2
Length=500
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 40/99 (40%), Positives = 57/99 (57%), Gaps = 0/99 (0%)
Query 48 GLSDDMCYHEFCRLIGKVNTSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHY 107
GLSD YHEFCRL+ ++ +++ L EL A + E Q + FT+ESL W PNS HY
Sbjct 243 GLSDPDNYHEFCRLLARLKSNYQLGELIAVPCYPEAIQLIAKFTVESLHLWLFAPNSVHY 302
Query 108 LLGVWAHMISPLLFYKQRVPKDLELYIERITAAFIMSRM 146
LL +W M++ + + K P L Y + A+I SR+
Sbjct 303 LLTLWQRMVASVPYVKSPDPHLLGTYTPEVIKAYIESRL 341
> Hs22056548_1
Length=583
Score = 31.2 bits (69), Expect = 0.98, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 29/54 (53%), Gaps = 6/54 (11%)
Query 70 HLSEL-CASEPFREFPQHLFDFTMESLK-SWQRMPNSK----HYLLGVWAHMIS 117
H+ +L AS P + Q+LF FT+E + +W R P ++ L G+W +S
Sbjct 431 HIHQLPAASNPVDQASQYLFAFTLEGQQFTWTRWPRAQGTTAASLRGLWPEKLS 484
> At5g55560
Length=313
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 30/60 (50%), Gaps = 6/60 (10%)
Query 66 NTSHHLSELCASEPFREFPQHLFDFTMESLKSWQRMPNSKHYLLGV-WAHMISPLLFYKQ 124
NT + ++E+C S RE+ + +M +LK W SK L G+ + H P + ++
Sbjct 104 NTLNFITEICTSGNLREYRKKHRHVSMRALKKW-----SKQILKGLDYLHTHDPCIIHRD 158
> At3g57060
Length=1439
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query 21 FFETTAIRNKYMGELILGTFQIIKTRTGLSDDMCYHEFCRLIGKVNTS-HHLSELCAS 77
F ++ + Y+ ++ +F + + T L D CR+IG T H++ + CAS
Sbjct 191 LFGSSDLDENYLSFIVKNSFTLFENATILKDAETKDALCRIIGASATKYHYIVQSCAS 248
Lambda K H
0.327 0.138 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2490594054
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40