bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1008_orf1
Length=137
Score E
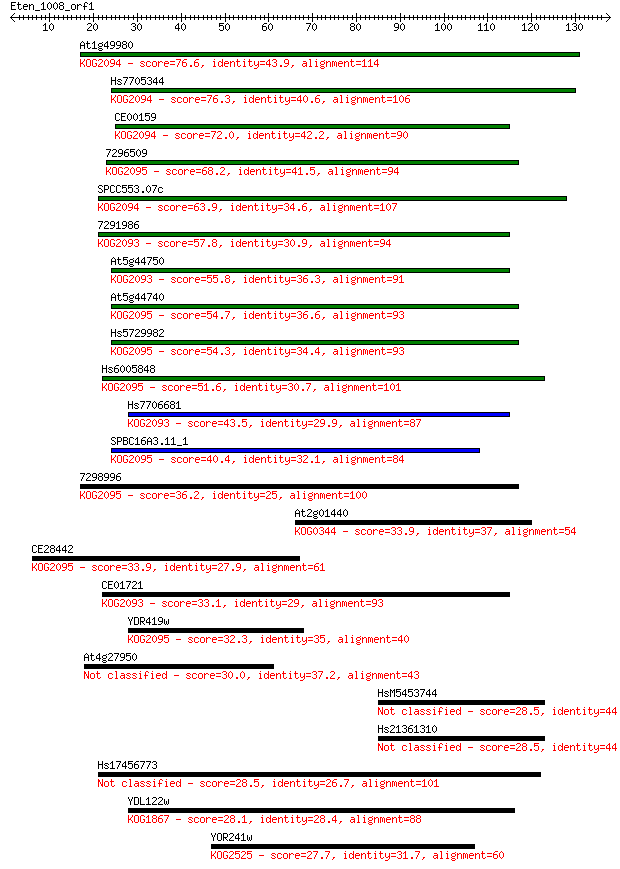
Sequences producing significant alignments: (Bits) Value
At1g49980 76.6 1e-14
Hs7705344 76.3 2e-14
CE00159 72.0 3e-13
7296509 68.2 5e-12
SPCC553.07c 63.9 7e-11
7291986 57.8 5e-09
At5g44750 55.8 2e-08
At5g44740 54.7 5e-08
Hs5729982 54.3 6e-08
Hs6005848 51.6 4e-07
Hs7706681 43.5 1e-04
SPBC16A3.11_1 40.4 0.001
7298996 36.2 0.017
At2g01440 33.9 0.083
CE28442 33.9 0.084
CE01721 33.1 0.16
YDR419w 32.3 0.27
At4g27950 30.0 1.3
HsM5453744 28.5 3.8
Hs21361310 28.5 3.8
Hs17456773 28.5 4.2
YDL122w 28.1 5.1
YOR241w 27.7 6.2
> At1g49980
Length=795
Score = 76.6 bits (187), Expect = 1e-14, Method: Composition-based stats.
Identities = 50/139 (35%), Positives = 73/139 (52%), Gaps = 32/139 (23%)
Query 17 CRRRVTA---LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATT 73
CR R + + E+R++V S TGL+CSAG A+R++AK+ SD+NKPNGQ +++ +T
Sbjct 210 CRERGLSGGEIAEELRSSVYSETGLTCSAGVAANRLLAKVCSDINKPNGQFVLQNDRST- 268
Query 74 AAATAATAAFLDSLNVRK---------------------IPGVGKYREKML-NALGIHTC 111
F+ L VRK I G+GK E +L +ALGI TC
Sbjct 269 ------VMTFVSFLPVRKEMVLCFYNLLVKLMPVGNGLQIGGIGKVTEHILKDALGIKTC 322
Query 112 GELLQHAPLLLYLLPKITA 130
E++Q LL L + +A
Sbjct 323 EEMVQKGSLLYALFSQSSA 341
> Hs7705344
Length=870
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 43/106 (40%), Positives = 57/106 (53%), Gaps = 7/106 (6%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V EIR + T L+ SAG + ++AK+ SD NKPNGQ + A F
Sbjct 293 VVKEIRFRIEQKTTLTASAGIAPNTMLAKVCSDKNKPNGQYQILPN-------RQAVMDF 345
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKIT 129
+ L +RK+ G+GK EKML ALGI TC EL Q LL L + +
Sbjct 346 IKDLPIRKVSGIGKVTEKMLKALGIITCTELYQQRALLSLLFSETS 391
> CE00159
Length=518
Score = 72.0 bits (175), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/90 (42%), Positives = 53/90 (58%), Gaps = 7/90 (7%)
Query 25 VSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFL 84
V EIR V TGL+CSAG ++ ++AKI SD+NKPNGQ ++E A FL
Sbjct 256 VREIRFRVEQLTGLTCSAGIASNFMLAKICSDLNKPNGQYVLEND-------KNAIMEFL 308
Query 85 DSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
L +RK+ G+G+ E L A+ I T G++
Sbjct 309 KDLPIRKVGGIGRVCEAQLKAMDIQTVGDM 338
> 7296509
Length=885
Score = 68.2 bits (165), Expect = 5e-12, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 56/95 (58%), Gaps = 10/95 (10%)
Query 23 ALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAA 82
++ E+RAAV TG CSAG ++I+AK+A+ MNKPN Q I+ T TA+
Sbjct 210 SVAGEVRAAVKKETGYECSAGIAHNKILAKLAAGMNKPNKQTILPLT---------ETAS 260
Query 83 FLDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
DSL V KI G+ GK+ E + LGI G++++
Sbjct 261 LFDSLPVGKIKGLGGKFGEVVCETLGIKFMGQVVK 295
> SPCC553.07c
Length=493
Score = 63.9 bits (154), Expect = 7e-11, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 62/108 (57%), Gaps = 8/108 (7%)
Query 21 VTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAAT 80
+T +V +IR V TG++ S G A++++AKIAS+ KPN Q +
Sbjct 248 ITMVVEKIRKQVHEETGVTVSCGIAANKLLAKIASNKRKPNNQFFIPFD-------EIGI 300
Query 81 AAFLDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYL-LPK 127
+ F++ L VR++ G+G+ E+ L L I TCG++ ++ +L Y+ LPK
Sbjct 301 SKFMNDLPVREVSGIGRVLEQQLLGLEIKTCGDIQRNLVILSYIFLPK 348
> 7291986
Length=995
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 52/94 (55%), Gaps = 8/94 (8%)
Query 21 VTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAAT 80
V A VS +R V S TG CSAG ++++A++A+ KPNGQ +++++
Sbjct 438 VMAFVSHLRQEVYSKTGCPCSAGVAGNKLLARMATKEAKPNGQFLLDSSNDIL------- 490
Query 81 AAFLDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
A++ +++ +PGVG L G++ CG++
Sbjct 491 -AYMAPMSLDLLPGVGSSISHKLKQAGLNNCGDV 523
> At5g44750
Length=955
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 46/91 (50%), Gaps = 9/91 (9%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
L S IR + TG S SAG G + ++A++A+ + KP GQ + A F
Sbjct 424 LASTIRNEILETTGCSASAGIGGTMLMARLATRVAKPAGQLYISA---------EKVEEF 474
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
LD L V +PGVG ++ L I TCG+L
Sbjct 475 LDQLPVGTLPGVGSVLKEKLVKQNIQTCGQL 505
> At5g44740
Length=689
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 51/94 (54%), Gaps = 10/94 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V+E+R V T +CSAG ++++AK+AS MNKP Q +V AA
Sbjct 203 IVAELRKQVLKETEFTCSAGIAHNKMLAKLASGMNKPAQQTVV---------PYAAVQEL 253
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
L SL ++K+ + GK + LG+ T G+LLQ
Sbjct 254 LSSLPIKKMKQLGGKLGTSLQTDLGVDTVGDLLQ 287
> Hs5729982
Length=713
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 49/94 (52%), Gaps = 10/94 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V E+RAA+ TG CSAG ++++AK+A +NKPN Q +V + +
Sbjct 196 IVEEMRAAIERETGFQCSAGISHNKVLAKLACGLNKPNRQTLV---------SHGSVPQL 246
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
+ +RKI + GK ++ LGI GEL Q
Sbjct 247 FSQMPIRKIRSLGGKLGASVIEILGIEYMGELTQ 280
> Hs6005848
Length=715
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 56/102 (54%), Gaps = 10/102 (9%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
+ + +E+R A+ + GL+ AG +++++AK+ S + KPN Q T +
Sbjct 177 SQIAAEMREAMYNQLGLTGCAGVASNKLLAKLVSGVFKPNQQ---------TVLLPESCQ 227
Query 82 AFLDSLN-VRKIPGVGKYREKMLNALGIHTCGELLQHAPLLL 122
+ SLN +++IPG+G K L ALGI++ +L +P +L
Sbjct 228 HLIHSLNHIKEIPGIGYKTAKCLEALGINSVRDLQTFSPKIL 269
> Hs7706681
Length=1251
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 9/87 (10%)
Query 28 IRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSL 87
+R + T + S G G++ ++A++A+ KP+GQ ++ F+
Sbjct 594 VRMEIKDQTKCAASVGIGSNILLARMATRKAKPDGQYHLKPEEVDD---------FIRGQ 644
Query 88 NVRKIPGVGKYREKMLNALGIHTCGEL 114
V +PGVG E L +LGI TCG+L
Sbjct 645 LVTNLPGVGHSMESKLASLGIKTCGDL 671
> SPBC16A3.11_1
Length=634
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 43/85 (50%), Gaps = 10/85 (11%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V EIR + +CSAG + +++K+ S NKPN Q I+ A +
Sbjct 222 IVKEIRDDIYLQLKYTCSAGVSFNPMLSKLVSSRNKPNKQTILTKN---------AIQDY 272
Query 84 LDSLNVRKIPGV-GKYREKMLNALG 107
L SL + I + GK+ E+++N LG
Sbjct 273 LVSLKITDIRMLGGKFGEEIINLLG 297
> 7298996
Length=737
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 54/104 (51%), Gaps = 13/104 (12%)
Query 17 CRRRV---TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATT 73
C +R+ T + EIR + G++C AG ++++AK+ ++PN Q ++ +T A
Sbjct 157 CAQRLAIGTRIAQEIREELKLRLGITCCAGIAYNKLLAKLVGSSHEPNQQTVLVSTYA-- 214
Query 74 AAATAATAAFLDSL-NVRKIPGVGKYREKMLNALGIHTCGELLQ 116
F+ L +++++ G+G+ + +L G+ + +L Q
Sbjct 215 -------EQFMRELGDLKRVTGIGQKTQCLLLEAGMSSVEQLQQ 251
> At2g01440
Length=845
Score = 33.9 bits (76), Expect = 0.083, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 5/54 (9%)
Query 66 VEATGATTAAATAATAAFLDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAP 119
VEAT AA A +DS+ PG+ K L++ G HT +LL H P
Sbjct 119 VEATSDDVFAAQRFLATSIDSM-----PGLSKRHSNQLDSCGFHTMKKLLHHFP 167
> CE28442
Length=603
Score = 33.9 bits (76), Expect = 0.084, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 6 LLQQKVSVRRECRRRVTALVS-----EIRAAVSSATGLSCSAGAGASRIVAKIASDMNKP 60
+L + + R C + L++ +IR + T CSAG G ++++AK+ +KP
Sbjct 172 VLLEYIENARNCTENLLLLIAAVTVEQIRQQIHEETQFFCSAGVGNNKMMAKLVCARHKP 231
Query 61 NGQKIV 66
Q ++
Sbjct 232 RQQTLI 237
> CE01721
Length=1027
Score = 33.1 bits (74), Expect = 0.16, Method: Composition-based stats.
Identities = 27/98 (27%), Positives = 41/98 (41%), Gaps = 17/98 (17%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
T L IR + T S G G++ ++A++A+ KP+G V A
Sbjct 562 TILAEHIRKVIREKTQCPASVGIGSTSLLARLATRHAKPDGVFWVNAHKKN--------- 612
Query 82 AFLDSLNVRKIPGVGKYREKMLNAL-----GIHTCGEL 114
F+ V+ +PG G +M+N L I C EL
Sbjct 613 EFISEEKVKDLPGFGY---EMMNRLTSFFGDITKCREL 647
> YDR419w
Length=632
Score = 32.3 bits (72), Expect = 0.27, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 28 IRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVE 67
IR ++ G + S G +++ V K+AS+ KP+ Q IV+
Sbjct 248 IRDSIKDILGYTTSCGLSSTKNVCKLASNYKKPDAQTIVK 287
> At4g27950
Length=334
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 18 RRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKP 60
RRRV L++EIR SS++ SA R + S + KP
Sbjct 65 RRRVKRLINEIRVEPSSSSTGDVSASPTKDRKRINVDSTVQKP 107
> HsM5453744
Length=444
Score = 28.5 bits (62), Expect = 3.8, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 85 DSLNVRKIPGVGKYREKMLNALGIHT-CGELLQHAPLLL 122
DSL+ RKI GV + R+ +L+AL + G L+Q +LL
Sbjct 153 DSLDERKIKGVIELRKSLLSALRTYEPYGSLVQQIRILL 191
> Hs21361310
Length=444
Score = 28.5 bits (62), Expect = 3.8, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 85 DSLNVRKIPGVGKYREKMLNALGIHT-CGELLQHAPLLL 122
DSL+ RKI GV + R+ +L+AL + G L+Q +LL
Sbjct 153 DSLDERKIKGVIELRKSLLSALRTYEPYGSLVQQIRILL 191
> Hs17456773
Length=158
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 54/115 (46%), Gaps = 14/115 (12%)
Query 21 VTALVSEIRAAVSSATGLSCSAGA-------GASRIVAKIASDMNK--PNGQKIVEATGA 71
++ L + +R V +ATG+S +A G + + + + D+ K + + G
Sbjct 17 LSYLPTMLRIFVFNATGISSNARFAQEFFIHGFTDMESSVLLDVRKLACSDNTVNFFYGF 76
Query 72 TTAAATAATAAFLDS---LNVRKIPGVGKYREKM--LNALGIHTCGELLQHAPLL 121
A + + F+ L ++ I G+G +RE++ LN H C L+ +AP++
Sbjct 77 FLALCMMSESVFITVSYVLILKTIMGIGSHRERLKALNTCVSHICAVLIFYAPVI 131
> YDL122w
Length=809
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 44/91 (48%), Gaps = 6/91 (6%)
Query 28 IRAAVSSATGLSC-SAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDS 86
IR +V S L+ + G++ ++++ SD +KP +I+E A TAA +
Sbjct 346 IRYSVFSGLSLNLPNENIGSTLKLSQLLSDWSKP---EIIEGVECNRCALTAAHSHLFGQ 402
Query 87 LNVRKIPGVGKYREKMLNALG--IHTCGELL 115
L + G EK++NA+ +H E+L
Sbjct 403 LKEFEKKPEGSIPEKLINAVKDRVHQIEEVL 433
> YOR241w
Length=548
Score = 27.7 bits (60), Expect = 6.2, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 47 SRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSLNVRKIPGVGKYREKMLNAL 106
SR + ASD NK N IV TG +TAA + + ++P +G Y L ++
Sbjct 107 SRRIGYSASDFNKLN---IVHITGTKGKGSTAAFTSSILGQYKEQLPRIGLYTSPHLKSV 163
Lambda K H
0.321 0.132 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40