bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1002_orf1
Length=300
Score E
Sequences producing significant alignments: (Bits) Value
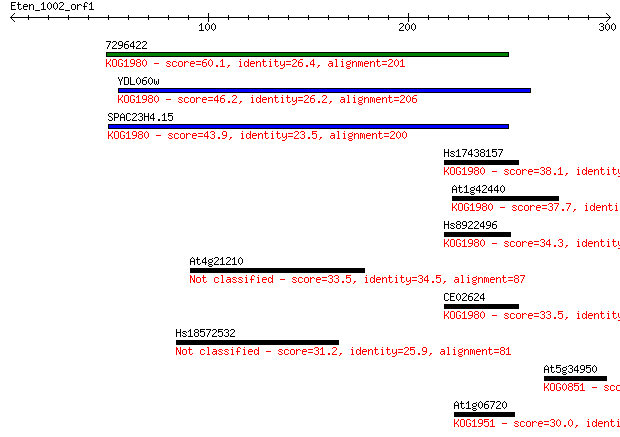
7296422 60.1 6e-09
YDL060w 46.2 7e-05
SPAC23H4.15 43.9 4e-04
Hs17438157 38.1 0.024
At1g42440 37.7 0.031
Hs8922496 34.3 0.34
At4g21210 33.5 0.60
CE02624 33.5 0.61
Hs18572532 31.2 2.9
At5g34950 30.8 4.1
At1g06720 30.0 6.1
> 7296422
Length=814
Score = 60.1 bits (144), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 53/204 (25%), Positives = 82/204 (40%), Gaps = 36/204 (17%)
Query 49 PPERPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVG 108
P R +EL+ +D CD + L + + F R G +I + QG+P V V
Sbjct 140 PVGRGNELI-ALDYLKVCDTTLLLTTAAFG-DDEIFDRWGQRIFNMMSAQGIPTPV--VA 195
Query 109 CVDPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAA 168
+D + P P K +A + + S+L E K + + ++ NV+R +G
Sbjct 196 LMDLESINPKRRPAAKQAAQKVI--------SKLLPEEKIMQLDTASEALNVMRRIGGQ- 246
Query 169 SLTQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEP---CLEACGLV 225
+R H +A H+ V P + P LE G +
Sbjct 247 --------------------KKRILHNVANRPHLFGDVVEFKPGSDPSDDLGTLEVTGFL 286
Query 226 RGAGLTCNLPVHITGVGDFVLSRI 249
RG L N VHI G+GDF LS++
Sbjct 287 RGQSLNVNGLVHIPGLGDFQLSQV 310
> YDL060w
Length=788
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 54/210 (25%), Positives = 93/210 (44%), Gaps = 37/210 (17%)
Query 55 ELLRYMDLCNCCDVL-VCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPS 113
++ ++++ +C V +FG S E G +I++AL+LQG+ S IGV +
Sbjct 141 DMTNFLNILDCAKVADFVVFGLSGVQEVD--EEFGEQIIRALELQGIA-SYIGVISNLSA 197
Query 114 VLEPADAPKGKSSASESLK-FMRRFFESELGAERKFFSIGSDADWHNVLRAV--GAAASL 170
V E K + +SL+ + + FF SE + +++ ++D NVLR + S+
Sbjct 198 VHE---KEKFQLDVKQSLESYFKHFFPSE----ERVYNLEKNSDALNVLRTLCQRLPRSI 250
Query 171 TQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGL 230
D RG+++A + + S D L G VRG G
Sbjct 251 NWRDN----------------RGYVVAD--FVDFVETSPDSGD-----LVIEGTVRGIGF 287
Query 231 TCNLPVHITGVGDFVLSRIETMEPAYEAKR 260
N VHI GDF L++IE + + + ++
Sbjct 288 NANRLVHIPDFGDFQLNKIEKISESSQKRK 317
> SPAC23H4.15
Length=783
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 47/200 (23%), Positives = 77/200 (38%), Gaps = 41/200 (20%)
Query 50 PERPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGC 109
P+R E +D C D ++ F S E F G I++ + QG+ + V
Sbjct 138 PKR--EFYSLIDACKVSDYVI--FVLSAVQEVDEF---GELIVRTTQGQGISSVLSMVHD 190
Query 110 VDPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAAS 169
L D+ K ++ +SL+ FF S+ + + F+ D NV+RA+ +
Sbjct 191 -----LSEVDSLKTRNEVKKSLQSFMNFFFSD---QERVFAADVSQDALNVMRALCTS-- 240
Query 170 LTQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAG 229
H + W + ++ + L G+VRG G
Sbjct 241 ------------------------HPRGIHWRDSRSYLLSQEISYSNGNLLVRGIVRGKG 276
Query 230 LTCNLPVHITGVGDFVLSRI 249
L N +HI G GDF ++RI
Sbjct 277 LDPNRLIHIQGFGDFAINRI 296
> Hs17438157
Length=465
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 218 CLEACGLVRGAGLTCNLPVHITGVGDFVLSRIET-MEP 254
LE G VRG L N +HITG GDF + I+ M+P
Sbjct 161 TLEISGYVRGQTLNVNRLLHITGRGDFQMKHIDDPMDP 198
> At1g42440
Length=766
Score = 37.7 bits (86), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 222 CGLVRGAGLTCNLPVHITGVGDFVLSRIETME---PAYEAKRREPLLVNGEAXAEV 274
G +R L+ N VH++GVGDF S+IE ++ P E K + + ++ EV
Sbjct 248 SGYLRARKLSVNQLVHVSGVGDFQFSKIEVLKDPFPLNERKNQNSMELDDSHDEEV 303
> Hs8922496
Length=656
Score = 34.3 bits (77), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 218 CLEACGLVRGAGLTCNLPVHITGVGDFVLSRIE 250
L+ G VRG L N +HI G GDF + +I+
Sbjct 123 TLKISGYVRGQTLNVNRLLHIVGYGDFQMKQID 155
> At4g21210
Length=403
Score = 33.5 bits (75), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 37/91 (40%), Gaps = 19/91 (20%)
Query 91 ILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFS 150
+L A+KLQG PP + S L P P G S S SE G+ERK
Sbjct 3 LLSAMKLQGRPPPI-------SSNLNPNSKPAGSDSVS--------LNASEPGSERKPRK 47
Query 151 IGSDADWHNVLRAVGAAASL----TQGDTNT 177
S + N R + + A L T G NT
Sbjct 48 FSSQLNRWNRARTLRSGAKLDSTITNGSNNT 78
> CE02624
Length=785
Score = 33.5 bits (75), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 218 CLEACGLVRGAGLTCNLPVHITGVGDFVLSRIE-TMEP 254
L A G +RG N VH+ G GDF +S+IE T++P
Sbjct 270 TLVAQGYLRGPEWNANNLVHLPGFGDFQISKIESTVDP 307
> Hs18572532
Length=188
Score = 31.2 bits (69), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 36/81 (44%), Gaps = 5/81 (6%)
Query 84 FSRRGYKILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGKSSASESLKFMRRFFESELG 143
++RG QAL +G P + P +EPA A K ++ E L +R + +
Sbjct 56 MAKRGQDTAQALASEGASPKSWHL----PRSVEPAGAQKSRTEVWEPLPRFQRMYGNAC- 110
Query 144 AERKFFSIGSDADWHNVLRAV 164
R+ F+ G W ++A+
Sbjct 111 MSRQKFATGVRPSWRTSVKAM 131
> At5g34950
Length=234
Score = 30.8 bits (68), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 268 GEAXAEVEAHVGNLQEVVGVVXTLCSFGYCC 298
E AE E HV + V+ V T+ SF YCC
Sbjct 122 DEVNAESEDHVVDAIGVITTVSTMSSFPYCC 152
> At1g06720
Length=1147
Score = 30.0 bits (66), Expect = 6.1, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 223 GLVRGAGLTCNLPVHITGVGDFVLSRIETM 252
G +RG + VHI GVGDF+++ + +
Sbjct 285 GYLRGCNFKKRMKVHIAGVGDFIVAGVTAL 314
Lambda K H
0.320 0.136 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6778584192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40