bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1001_orf3
Length=654
Score E
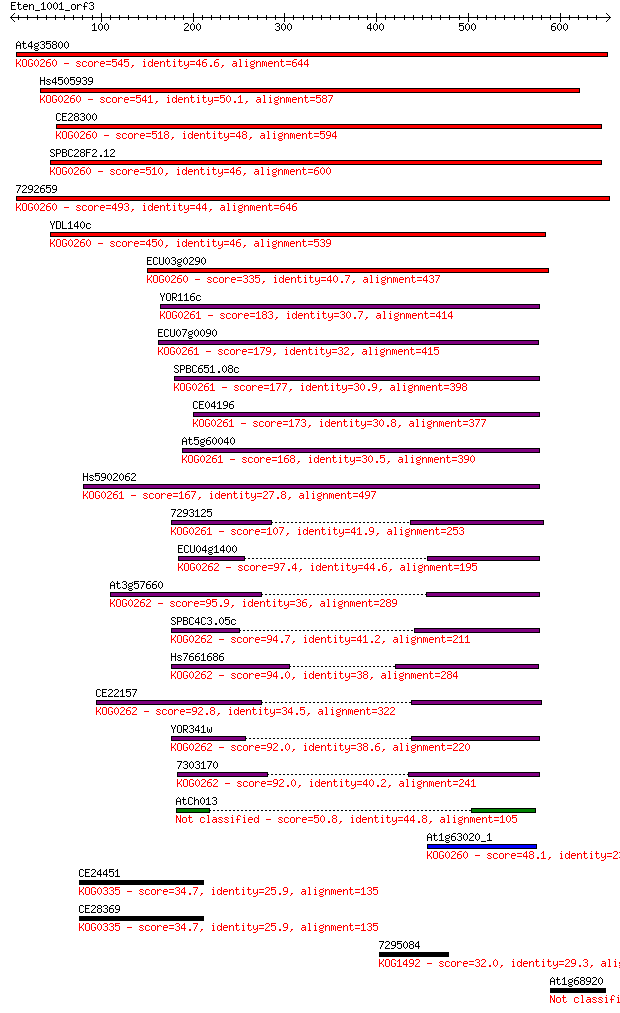
Sequences producing significant alignments: (Bits) Value
At4g35800 545 1e-154
Hs4505939 541 1e-153
CE28300 518 1e-146
SPBC28F2.12 510 3e-144
7292659 493 5e-139
YDL140c 450 4e-126
ECU03g0290 335 2e-91
YOR116c 183 1e-45
ECU07g0090 179 2e-44
SPBC651.08c 177 5e-44
CE04196 173 9e-43
At5g60040 168 4e-41
Hs5902062 167 8e-41
7293125 107 1e-22
ECU04g1400 97.4 1e-19
At3g57660 95.9 2e-19
SPBC4C3.05c 94.7 5e-19
Hs7661686 94.0 1e-18
CE22157 92.8 2e-18
YOR341w 92.0 3e-18
7303170 92.0 4e-18
AtCh013 50.8 9e-06
At1g63020_1 48.1 6e-05
CE24451 34.7 0.77
CE28369 34.7 0.79
7295084 32.0 4.9
At1g68920 30.8 9.9
> At4g35800
Length=1840
Score = 545 bits (1404), Expect = 1e-154, Method: Compositional matrix adjust.
Identities = 300/649 (46%), Positives = 423/649 (65%), Gaps = 35/649 (5%)
Query 8 MQMDSEKIRRVYKHHFDQESYGKGWILDEELRTRLSVDFAAQQVLEDEFETIKEDKLRLC 67
++M + R +K+ D E++ ++ DE L + + V + E+ ++ D+ +L
Sbjct 906 LKMKKSEFDRTFKYEIDDENWNPTYLSDEHLEDLKGIR-ELRDVFDAEYSKLETDRFQLG 964
Query 68 KTIFPDGEVKQHIPINIMRLVEFAKAQFPASVEQLRKQDPVAIAKSVQELLNEKLVVVKQ 127
I +G+ +P+NI R + A+ F + ++ PV I +V +L E+L+VV
Sbjct 965 TEIATNGDSTWPLPVNIKRHIWNAQKTFKIDLRKISDMHPVEIVDAVDKL-QERLLVVPG 1023
Query 128 TCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQLGAKAVHWLLGEVERQFHRSLAH 187
D +S E Q+NAT+F LR+ L S+R+LE +L +A W++GE+E +F +SL
Sbjct 1024 D---DALSVEAQKNATLFFNILLRSTLASKRVLEEYKLSREAFEWVIGEIESRFLQSLVA 1080
Query 188 AGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVY 247
GE +G +AAQSIGEPATQMTLNTFH+AGV +KNVTLGVPRL+E+INVAK +KTPSL+VY
Sbjct 1081 PGEMIGCVAAQSIGEPATQMTLNTFHYAGVSAKNVTLGVPRLREIINVAKRIKTPSLSVY 1140
Query 248 LLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDD 307
L E + +E AK VQ LE+TTL V +V YDPDP TI+ +D ++V YYE PD+
Sbjct 1141 LTPEASKSKEGAKTVQCALEYTTLRSVTQATEVWYDPDPMSTIIEEDFEFVRSYYEMPDE 1200
Query 308 EELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDNSDELVLRI 367
+ P + WLLR++L+ ++M+DKKL M +I E I EFD D+L CI+ DDN+ +L+LRI
Sbjct 1201 DVSPDKISPWLLRIELNREMMVDKKLSMADIAEKINLEFD-DDLTCIFNDDNAQKLILRI 1259
Query 368 RIKKHGQEQDPNSD--DQSSEIENRFLQKLMLHCLVGITLRGVSRITKVYMREEARTLYN 425
RI ++ P + D+S+E ++ FL+K+ + L + LRG+ I KV++++ ++ ++
Sbjct 1260 RIM---NDEGPKGELQDESAE-DDVFLKKIESNMLTEMALRGIPDINKVFIKQVRKSRFD 1315
Query 426 ETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLREL 485
E G F+ W+LDT+G NL V+ VD RTTSN + EI +VLGIEAVRRALL EL
Sbjct 1316 E-EGGFKTSEEWMLDTEGVNLLAVMCHEDVDPKRTTSNHLIEIIEVLGIEAVRRALLDEL 1374
Query 486 RAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEA 545
R VISFDGSYVNYRHLAILCDTMT +G+LM+ITRHGINR + GPL +CSFEETV+IL++A
Sbjct 1375 RVVISFDGSYVNYRHLAILCDTMTYRGHLMAITRHGINRNDTGPLMRCSFEETVDILLDA 1434
Query 546 AVFAETDHLRGVTENIMLGQLCPLGTGDVDLLIDEEKLKDAHRAVEPQQDLALEGLASPG 605
A +AETD LRGVTENIMLGQL P+GTGD +L +++E LK+ A+E Q ++GL
Sbjct 1435 AAYAETDCLRGVTENIMLGQLAPIGTGDCELYLNDEMLKN---AIELQLPSYMDGLEF-- 1489
Query 606 GADGMSPDG---SGTPSPHNVTSVFSPMPFSTVYSSFAQSPSNPLSPTS 651
GM+P SGTP + S ++ SP+ LSP S
Sbjct 1490 ---GMTPARSPVSGTPYHEGMMS-----------PNYLLSPNMRLSPMS 1524
> Hs4505939
Length=1970
Score = 541 bits (1395), Expect = 1e-153, Method: Compositional matrix adjust.
Identities = 294/597 (49%), Positives = 396/597 (66%), Gaps = 25/597 (4%)
Query 34 LDEELRTRLSVDFAAQQVLEDEFETIKEDKLRLCKTIFPDGEVKQHIPINIMRLVEFAKA 93
L E+L + + Q LE EFE ++ED+ + + IFP G+ K +P N++R++ A+
Sbjct 934 LQEDLVKDVLSNAHIQNELEREFERMREDR-EVLRVIFPTGDSKVVLPCNLLRMIWNAQK 992
Query 94 QFPASVEQLRKQDPVAIAKSVQELLNEKLVVVKQTCKADTISAEVQENATIFMKAHLRTV 153
F + P+ + + V+EL ++KLV+V D +S + QENAT+ HLR+
Sbjct 993 IFHINPRLPSDLHPIKVVEGVKEL-SKKLVIVNGD---DPLSRQAQENATLLFNIHLRST 1048
Query 154 LNSRRLLEREQLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFH 213
L SRR+ E +L +A WLLGE+E +F++++AH GE VGA+AAQS+GEPATQMTLNTFH
Sbjct 1049 LCSRRMAEEFRLSGEAFDWLLGEIESKFNQAIAHPGEMVGALAAQSLGEPATQMTLNTFH 1108
Query 214 FAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEK 273
+AGV +KNVTLGVPRLKELIN++K KTPSLTV+LL + A D ERAKD+ LEHTTL K
Sbjct 1109 YAGVSAKNVTLGVPRLKELINISKKPKTPSLTVFLLGQSARDAERAKDILCRLEHTTLRK 1168
Query 274 VCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGRWLLRVQLDSKVMIDKKL 333
V + YDP+P T+V +D++WV+ YYE PD + + WLLRV+LD K M D+KL
Sbjct 1169 VTANTAIYYDPNPQSTVVAEDQEWVNVYYEMPDFDV--ARISPWLLRVELDRKHMTDRKL 1226
Query 334 PMKEIGETIYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEIENRFLQ 393
M++I E I F D+L+CI+ DDN+++LVLRIRI D N + E+ ++
Sbjct 1227 TMEQIAEKINAGF-GDDLNCIFNDDNAEKLVLRIRI----MNSDENKMQEEEEVVDKMDD 1281
Query 394 KLMLHC-----LVGITLRGVSRITKVYMR----EEARTLYNETTGKFERVNNWVLDTDGC 444
+ L C L +TL+G+ +I+KVYM + + + G+F+ + W+L+TDG
Sbjct 1282 DVFLRCIESNMLTDMTLQGIEQISKVYMHLPQTDNKKKIIITEDGEFKALQEWILETDGV 1341
Query 445 NLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAIL 504
+L VL+ VD RTTSNDI EIF VLGIEAVR+AL REL VISFDGSYVNYRHLA+L
Sbjct 1342 SLMRVLSEKDVDPVRTTSNDIVEIFTVLGIEAVRKALERELYHVISFDGSYVNYRHLALL 1401
Query 505 CDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLG 564
CDTMT +G+LM+ITRHG+NR + GPL KCSFEETV++LMEAA E+D ++GV+ENIMLG
Sbjct 1402 CDTMTCRGHLMAITRHGVNRQDTGPLMKCSFEETVDVLMEAAAHGESDPMKGVSENIMLG 1461
Query 565 QLCPLGTGDVDLLIDEEKLKDAHRAVEPQQDLALEGLASPGGA-DGMSPDGSGTPSP 620
QL P GTG DLL+D EK K +E ++ G A P G G +P G SP
Sbjct 1462 QLAPAGTGCFDLLLDAEKCK---YGMEIPTNIPGLGAAGPTGMFFGSAPSPMGGISP 1515
> CE28300
Length=1852
Score = 518 bits (1335), Expect = 1e-146, Method: Compositional matrix adjust.
Identities = 285/604 (47%), Positives = 401/604 (66%), Gaps = 34/604 (5%)
Query 51 VLEDEFETIKEDKLRLCKTIFPDGEVKQHIPINIMRLVEFAKAQFPASVEQLRKQDPVAI 110
++E E+ ++ED+ RL + IFP G+ K +P N+ RL+ A+ F + + P+ +
Sbjct 941 LVESEWSQLEEDR-RLLRKIFPRGDAKIVLPCNLQRLIWNAQKIFKVDLRKPVNLSPLHV 999
Query 111 AKSVQELLNEKLVVVKQTCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQLGAKAV 170
V+EL ++KL++V D IS + Q NAT+ M LR+ L ++ + + +L ++A
Sbjct 1000 ISGVREL-SKKLIIVSGN---DEISKQAQYNATLLMNILLRSTLCTKNMCTKSKLNSEAF 1055
Query 171 HWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLK 230
WLLGE+E +F +++A GE VGA+AAQS+GEPATQMTLNTFH+AGV +KNVTLGVPRLK
Sbjct 1056 DWLLGEIESRFQQAIAQPGEMVGALAAQSLGEPATQMTLNTFHYAGVSAKNVTLGVPRLK 1115
Query 231 ELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTI 290
E+INV+KT+KTPSLTV+L A D E+AKDV LEHTTL+KV + YDPDP T+
Sbjct 1116 EIINVSKTLKTPSLTVFLTGAAAKDPEKAKDVLCKLEHTTLKKVTCNTAIYYDPDPKNTV 1175
Query 291 VPQDKQWVSDYYEFPDDEELPQSLGR---WLLRVQLDSKVMIDKKLPMKEIGETIYREFD 347
+ +D++WVS +YE PD + L R WLLR++LD K M+DKKL M+ I + I+ F
Sbjct 1176 IAEDEEWVSIFYEMPDHD-----LSRTSPWLLRIELDRKRMVDKKLTMEMIADRIHGGFG 1230
Query 348 NDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEIENR-FLQKLMLHCLVGITLR 406
ND + I+TDDN+++LV R+RI G+++ ++Q ++E+ FL+ + + L +TL+
Sbjct 1231 ND-VHTIYTDDNAEKLVFRLRIA--GEDKGEAQEEQVDKMEDDVFLRCIEANMLSDLTLQ 1287
Query 407 GVSRITKVYMRE----EARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTS 462
G+ I+KVYM + + + + G F+ V +W+L+TDG L VL+ ++D RTTS
Sbjct 1288 GIPAISKVYMNQPNTDDKKRIIITPEGGFKSVADWILETDGTALLRVLSERQIDPVRTTS 1347
Query 463 NDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGI 522
NDI EIF+VLGIEAVR+A+ RE+ VISFDGSYVNYRHLA+LCD MT KG+LM+ITRHGI
Sbjct 1348 NDICEIFEVLGIEAVRKAIEREMDNVISFDGSYVNYRHLALLCDVMTAKGHLMAITRHGI 1407
Query 523 NRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLLIDEEK 582
NR E G L +CSFEETV+ILMEAAV AE D ++GV+ENIMLGQL GTG DL++D EK
Sbjct 1408 NRQEVGALMRCSFEETVDILMEAAVHAEEDPVKGVSENIMLGQLARCGTGCFDLVLDVEK 1467
Query 583 LKDAHRAVEPQQDLALEGLASPGGADGMSPDGSGTPSPHNVTSVFSPM--PFSTVYSSFA 640
K +E Q++ + G G +G+PS + SP + Y+ A
Sbjct 1468 CK---YGMEIPQNVVMGG--------GFYGSFAGSPSNREFSPAHSPWNSGVTPTYAGAA 1516
Query 641 QSPS 644
SP+
Sbjct 1517 WSPT 1520
> SPBC28F2.12
Length=1752
Score = 510 bits (1314), Expect = 3e-144, Method: Compositional matrix adjust.
Identities = 276/606 (45%), Positives = 386/606 (63%), Gaps = 22/606 (3%)
Query 45 DFAAQQVLEDEFETIKEDKLRLCKTIFPDGEVKQHIPINIMRLVEFAKAQFPASVEQLRK 104
D + Q +L++E+ + D+ LCK IFP G+ + +P+N+ R+++ A F ++
Sbjct 924 DSSVQDLLDEEYTQLVADRELLCKFIFPKGDARWPLPVNVQRIIQNALQIFHLEAKKPTD 983
Query 105 QDPVAIAKSVQELLNEKLVVVKQTCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQ 164
P I + EL+ KL + + +D I+ +VQ NAT+ + LR+ +R++ +
Sbjct 984 LLPSDIINGLNELI-AKLTIFR---GSDRITRDVQNNATLLFQILLRSKFAVKRVIMEYR 1039
Query 165 LGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTL 224
L A W++GEVE +F +++ GE VG +AAQSIGEPATQMTLNTFH+AGV SKNVTL
Sbjct 1040 LNKVAFEWIMGEVEARFQQAVVSPGEMVGTLAAQSIGEPATQMTLNTFHYAGVSSKNVTL 1099
Query 225 GVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDP 284
GVPRLKE++NVAK +KTPSLT+YL+ A + + AK+VQT +EHTTL V + ++ YDP
Sbjct 1100 GVPRLKEILNVAKNIKTPSLTIYLMPWIAANMDLAKNVQTQIEHTTLSTVTSATEIHYDP 1159
Query 285 DPTKTIVPQDKQWVSDYYEFPDDEELPQSLGR---WLLRVQLDSKVMIDKKLPMKEIGET 341
DP T++ +DK +V ++ PD EE+ ++L + WLLR++LD M+DKKL M ++
Sbjct 1160 DPQDTVIEEDKDFVEAFFAIPD-EEVEENLYKQSPWLLRLELDRAKMLDKKLSMSDVAGK 1218
Query 342 IYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCLV 401
I F+ D L IW++DN+D+L++R RI + + + D+ E + FL+ + H L
Sbjct 1219 IAESFERD-LFTIWSEDNADKLIIRCRIIRDDDRKAEDDDNMIEE--DVFLKTIEGHMLE 1275
Query 402 GITLRGVSRITKVYMREEARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTT 461
I+LRGV IT+VYM E + + G FER + WVL+TDG NL E +T VD+TRT
Sbjct 1276 SISLRGVPNITRVYMMEH-KIVRQIEDGTFERADEWVLETDGINLTEAMTVEGVDATRTY 1334
Query 462 SNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHG 521
SN EI +LGIEA R ALL+ELR VI FDGSYVNYRHLA+LCD MT +G+LM+ITRHG
Sbjct 1335 SNSFVEILQILGIEATRSALLKELRNVIEFDGSYVNYRHLALLCDVMTSRGHLMAITRHG 1394
Query 522 INRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLLIDEE 581
INR E G L +CSFEETVEILM+AA E D +G++ENIMLGQL P+GTG D+ +D++
Sbjct 1395 INRAETGALMRCSFEETVEILMDAAASGEKDDCKGISENIMLGQLAPMGTGAFDIYLDQD 1454
Query 582 KLKDAHRAVEPQQDLALEGLASPGGADGMSPDGSGTP---SPHNVTSVFSPMPFSTVYSS 638
L + A+ LA G P+G+GTP SP V S F P + +S
Sbjct 1455 MLMNYSLGT------AVPTLAGSGMGTSQLPEGAGTPYERSPM-VDSGFVGSPDAAAFSP 1507
Query 639 FAQSPS 644
Q S
Sbjct 1508 LVQGGS 1513
> 7292659
Length=1887
Score = 493 bits (1269), Expect = 5e-139, Method: Compositional matrix adjust.
Identities = 284/653 (43%), Positives = 411/653 (62%), Gaps = 20/653 (3%)
Query 8 MQMDSEKIRRVYKHHFDQESYGKGWILDEELRTRLSVDFAAQQVLEDEFETIKEDKLRLC 67
+++ ++ + +K + E K D+ ++ ++ A Q LE E++ + D+ L
Sbjct 901 VKLSNKSFEKRFKFDWSNERLMKKVFTDDVIK-EMTDSSEAIQELEAEWDRLVSDRDSL- 958
Query 68 KTIFPDGEVKQHIPINIMRLVEFAKAQFPASVEQLRKQDPVAIAKSVQELLNEKLVVVKQ 127
+ IFP+GE K +P N+ R++ + F + P+ + K V+ LL E+ V+V
Sbjct 959 RQIFPNGESKVVLPCNLQRMIWNVQKIFHINKRLPTDLSPIRVIKGVKTLL-ERCVIVTG 1017
Query 128 TCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQLGAKAVHWLLGEVERQFHRSLAH 187
D IS + ENAT+ + +R+ L ++ + E +L +A WL+GE+E +F ++ A+
Sbjct 1018 N---DRISKQANENATLLFQCLIRSTLCTKYVSEEFRLSTEAFEWLVGEIETRFQQAQAN 1074
Query 188 AGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVY 247
GE VGA+AAQS+GEPATQMTLNTFHFAGV SKNVTLGVPRLKE+IN++K K PSLTV+
Sbjct 1075 PGEMVGALAAQSLGEPATQMTLNTFHFAGVSSKNVTLGVPRLKEIINISKKPKAPSLTVF 1134
Query 248 LLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDD 307
L A D E+AK+V LEHTTL KV + YDPDP +T++ +D+++V+ YYE PD
Sbjct 1135 LTGGAARDAEKAKNVLCRLEHTTLRKVTANTAIYYDPDPQRTVISEDQEFVNVYYEMPDF 1194
Query 308 EELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDNSDELVLRI 367
+ P + WLLR++LD K M DKKL M++I E I F D L+CI+ DDN+D+LVLRI
Sbjct 1195 D--PTRISPWLLRIELDRKRMTDKKLTMEQIAEKINVGFGED-LNCIFNDDNADKLVLRI 1251
Query 368 RIKKHGQEQDPNSDDQSSEIE-NRFLQKLMLHCLVGITLRGVSRITKVYMR-----EEAR 421
RI + + + + D+ ++E + FL+ + + L +TL+G+ I KVYM + R
Sbjct 1252 RIMNNEENKFQDEDEAVDKMEDDMFLRCIEANMLSDMTLQGIEAIGKVYMHLPQTDSKKR 1311
Query 422 TLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRAL 481
+ E TG+F+ + W+L+TDG ++ +VL+ VD RT+SNDI EIF VLGIEAVR+++
Sbjct 1312 IVITE-TGEFKAIGEWLLETDGTSMMKVLSERDVDPIRTSSNDICEIFQVLGIEAVRKSV 1370
Query 482 LRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEI 541
+E+ AV+ F G YVNYRHLA+LCD MT KG+LM+ITRHGINR + G L +CSFEETV++
Sbjct 1371 EKEMNAVLQFYGLYVNYRHLALLCDVMTAKGHLMAITRHGINRQDTGALMRCSFEETVDV 1430
Query 542 LMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLLIDEEKLKDAHRAVEPQQDLALEGL 601
LM+AA AETD +RGV+ENI++GQL +GTG DLL+D EK + + L G
Sbjct 1431 LMDAAAHAETDPMRGVSENIIMGQLPKMGTGCFDLLLDAEKCRFGIEIPNTLGNSMLGGA 1490
Query 602 ASPGGADGMSPDGSGTPSPHNVTSVFSPMPFSTVYSSFAQSPSNP-LSPTSPS 653
A G G +P S TP + +P FS A +P P SP++ S
Sbjct 1491 AMFIGG-GSTP--SMTPPMTPWANCNTPRYFSPPGHVSAMTPGGPSFSPSAAS 1540
> YDL140c
Length=1733
Score = 450 bits (1158), Expect = 4e-126, Method: Compositional matrix adjust.
Identities = 248/545 (45%), Positives = 351/545 (64%), Gaps = 22/545 (4%)
Query 45 DFAAQQVLEDEFETIKEDKLRLCKTIFPDGEVKQHIPINIMRLVEFAKAQFPASVEQLRK 104
D Q +L++E++ + +D+ + + +F DGE +P+NI R+++ A+ F ++ +
Sbjct 922 DLKLQVLLDEEYKQLVKDR-KFLREVFVDGEANWPLPVNIRRIIQNAQQTF--HIDHTKP 978
Query 105 QDPVA--IAKSVQELLNEKLVVVKQTCKADTISAEVQENATIFMKAHLRTVLNSRRLLER 162
D I V++L E L+V++ + I Q +A LR+ L +RR+L+
Sbjct 979 SDLTIKDIVLGVKDL-QENLLVLR---GKNEIIQNAQRDAVTLFCCLLRSRLATRRVLQE 1034
Query 163 EQLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNV 222
+L +A W+L +E QF RS+ H GE VG +AAQSIGEPATQMTLNTFHFAGV SK V
Sbjct 1035 YRLTKQAFDWVLSNIEAQFLRSVVHPGEMVGVLAAQSIGEPATQMTLNTFHFAGVASKKV 1094
Query 223 TLGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIY 282
T GVPRLKE++NVAK +KTPSLTVYL A DQE+AK +++ +EHTTL+ V +++ Y
Sbjct 1095 TSGVPRLKEILNVAKNMKTPSLTVYLEPGHAADQEQAKLIRSAIEHTTLKSVTIASEIYY 1154
Query 283 DPDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGR---WLLRVQLDSKVMIDKKLPMKEIG 339
DPDP T++P+D++ + ++ DEE QS + WLLR++LD M DK L M ++G
Sbjct 1155 DPDPRSTVIPEDEEIIQLHFSLL-DEEAEQSFDQQSPWLLRLELDRAAMNDKDLTMGQVG 1213
Query 340 ETIYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEI-ENRFLQKLMLH 398
E I + F ND L IW++DN ++L++R R+ + P S D +E E+ L+K+
Sbjct 1214 ERIKQTFKND-LFVIWSEDNDEKLIIRCRVVR------PKSLDAETEAEEDHMLKKIENT 1266
Query 399 CLVGITLRGVSRITKVYMREEARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDST 458
L ITLRGV I +V M + R + + TG++ + WVL+TDG NL EV+T P +D T
Sbjct 1267 MLENITLRGVENIERVVMMKYDRKVPS-PTGEYVKEPEWVLETDGVNLSEVMTVPGIDPT 1325
Query 459 RTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSIT 518
R +N +I +VLGIEA R AL +E+ VI+ DGSYVNYRH+A+L D MT +G L S+T
Sbjct 1326 RIYTNSFIDIMEVLGIEAGRAALYKEVYNVIASDGSYVNYRHMALLVDVMTTQGGLTSVT 1385
Query 519 RHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLLI 578
RHG NR G L +CSFEETVEIL EA AE D RGV+EN++LGQ+ P+GTG D++I
Sbjct 1386 RHGFNRSNTGALMRCSFEETVEILFEAGASAELDDCRGVSENVILGQMAPIGTGAFDVMI 1445
Query 579 DEEKL 583
DEE L
Sbjct 1446 DEESL 1450
> ECU03g0290
Length=1599
Score = 335 bits (860), Expect = 2e-91, Method: Compositional matrix adjust.
Identities = 178/437 (40%), Positives = 268/437 (61%), Gaps = 27/437 (6%)
Query 150 LRTVLNSRRLLEREQLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTL 209
+RT L+ +R+L +L +A +W++ ++ + ++ E VG +AAQS+GEPATQMTL
Sbjct 1007 IRTNLSVKRVLLEHRLNTEAFNWVVEVIDAKILKAKITPNEMVGTLAAQSVGEPATQMTL 1066
Query 210 NTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHT 269
NTFH AGV S VT+GVPRLKE+ NV K +KTPS+ +YL E E AK +Q +E
Sbjct 1067 NTFHLAGVAS-TVTMGVPRLKEIFNVTKNLKTPSMKIYLDREHGKSIEAAKTIQNEIECL 1125
Query 270 TLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGRWLLRVQLDSKVMI 329
T++ +C +++ YDP+ T T + DK +V Y+EFPD++ L +L+R+ +D ++
Sbjct 1126 TVKDLCLFSEIYYDPEITGTEISDDKDFVEAYFEFPDEDVDFSCLSPFLMRLVVDRAKLV 1185
Query 330 DKKLPMKEIGETIYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEIEN 389
+ + ++ + I +E I +D+N+ +V+R+R K SE E+
Sbjct 1186 GRGINLEYVAMFIRKELGGGA-HVICSDENAVNMVVRVRTTK-------------SEDES 1231
Query 390 RFLQKLMLHCLVGITLRGVSRITKVYMREEARTLYNETTGKFERVNNWVLDTDGCNLDEV 449
L+ L+ + L G + KVY+ E+ W L TDG L ++
Sbjct 1232 LNFYTTALNSLLRLQLGGYKNVKKVYISEDKDR------------KEWYLQTDGICLSQI 1279
Query 450 LTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMT 509
L P V+S T SND+ EI + LGIEA R ++LREL VI +GSYVNYRH+++L D MT
Sbjct 1280 LGNPAVNSRLTISNDLVEIAETLGIEAARESILRELTIVIDGNGSYVNYRHMSLLADVMT 1339
Query 510 QKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPL 569
+GYL ITRHG+N+V G LK+ SFEETVEIL++AA+ +E + RG+TENIM+GQL P+
Sbjct 1340 MRGYLCGITRHGVNKVGAGALKRSSFEETVEILLDAALVSEKNICRGITENIMMGQLAPM 1399
Query 570 GTGDVDLLIDEEKLKDA 586
GTG++++++D +KL A
Sbjct 1400 GTGNIEIMLDMKKLDKA 1416
> YOR116c
Length=1460
Score = 183 bits (465), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 127/419 (30%), Positives = 213/419 (50%), Gaps = 53/419 (12%)
Query 164 QLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVT 223
++ K+V L ++ ++ G +GAI AQSIGEP TQMTL TFHFAGV S NVT
Sbjct 1061 RISEKSVRKFLEIALFKYRKARLEPGTAIGAIGAQSIGEPGTQMTLKTFHFAGVASMNVT 1120
Query 224 LGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYD 283
LGVPR+KE+IN +K + TP + L+++ D+ A+ V+ +E T L V Q +Y
Sbjct 1121 LGVPRIKEIINASKVISTPIINAVLVNDN--DERAARVVKGRVEKTLLSDVAFYVQDVYK 1178
Query 284 PDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIY 343
+L +R+ L + + +L +++I I
Sbjct 1179 ----------------------------DNLSFIQVRIDLGTIDKLQLELTIEDIAVAIT 1210
Query 344 REFDNDELDCIWTDDN---SDELVLRIRIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCL 400
R +L +D N D + + + + + + S + SE + + + + L
Sbjct 1211 RA---SKLKIQASDVNIIGKDRIAINVFPEGYKAKSISTSAKEPSENDVFYRMQQLRRAL 1267
Query 401 VGITLRGVSRITK--VYMREEARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDST 458
+ ++G+ I++ + +R++ GK E L +G L +V+ V +
Sbjct 1268 PDVVVKGLPDISRAVINIRDD---------GKRE------LLVEGYGLRDVMCTDGVIGS 1312
Query 459 RTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSIT 518
RTT+N + E+F VLGIEA R +++RE+ +S G V+ RH+ +L D MT KG ++ IT
Sbjct 1313 RTTTNHVLEVFSVLGIEAARYSIIREINYTMSNHGMSVDPRHIQLLGDVMTYKGEVLGIT 1372
Query 519 RHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLL 577
R G++++ L+ SFE+T + L +AA + + D + GV+E I+LGQ +GTG ++
Sbjct 1373 RFGLSKMRDSVLQLASFEKTTDHLFDAAFYMKKDAVEGVSECIILGQTMSIGTGSFKVV 1431
> ECU07g0090
Length=1349
Score = 179 bits (453), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 133/415 (32%), Positives = 207/415 (49%), Gaps = 66/415 (15%)
Query 162 REQLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKN 221
R L + V ++ + G VGAIA QSIGEP TQMTL TFHFAGV S N
Sbjct 995 RYFLSKDFIETFFSMVSQKMMNLIVEPGTTVGAIAGQSIGEPGTQMTLKTFHFAGVASMN 1054
Query 222 VTLGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVI 281
+TLGVPRLKE+IN + TP + L D +D R + ++ L+ +L+ VC+
Sbjct 1055 ITLGVPRLKEIINAVCNISTPIINAELDD--PHDLFRTEVIKGRLDRISLKDVCS----- 1107
Query 282 YDPDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGET 341
++ +D+ ++ +RV L+S + L + +I +
Sbjct 1108 ----SLTEVISKDEIFLD-------------------IRVDLESLSRLKLNLDIHKIEKL 1144
Query 342 IYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCLV 401
+ ++D++ + +E LR+ IKK DQS R +KL+
Sbjct 1145 LN---NHDQVRVV------NENTLRVSIKK--------ITDQSYFNLQRIKKKLL----- 1182
Query 402 GITLRGVSRITKVYMREEARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTT 461
+ G ++ +V + +R LY+ L +G L V+ V + TT
Sbjct 1183 NTKICGAPQVNRVII-NSSRGLYS-------------LVIEGRGLLNVINIDGVKAINTT 1228
Query 462 SNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHG 521
SN ITE+ +VLGIEA R ++ E+ IS G ++ RH+ +L DTMT +G + ITR G
Sbjct 1229 SNSITEVEEVLGIEAARAQIIHEIEYTISNHGIKIDPRHIMLLADTMTYRGEVFGITRFG 1288
Query 522 INRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDL 576
I+++ + L SFE+T + L EAAV +++D + GV+E+I+LG +GTG +DL
Sbjct 1289 ISKMSRSTLMLASFEQTSDYLFEAAVQSKSDEICGVSESIILGIPICIGTGSMDL 1343
> SPBC651.08c
Length=1405
Score = 177 bits (450), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 123/398 (30%), Positives = 201/398 (50%), Gaps = 55/398 (13%)
Query 180 QFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTV 239
++ R+ G VGAI AQSIGEP TQMTL TFHFAGV ++ TLGVPR+KE+IN AKT+
Sbjct 1035 KYMRAKVEPGTAVGAIGAQSIGEPGTQMTLKTFHFAGVAAQT-TLGVPRIKEIINAAKTI 1093
Query 240 KTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVS 299
TP +T L+++ D+ A+ V+ +E T L+ V + + +Y P V+
Sbjct 1094 STPIITGQLIND--RDERSARVVKGRIEKTYLKDVTSYIEEVYGP-------------VT 1138
Query 300 DYYEFPDDEELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDN 359
Y ++V D+ + + + +I I+ + +L
Sbjct 1139 TYLS---------------IQVNFDTISKLQLDITLADIAAAIW---NTPKLKIPSQQVT 1180
Query 360 SDELVLRIRIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCLVGITLRGVSRITKVYMREE 419
+ + +I + +SD +SSE E + + L + + G+ I
Sbjct 1181 VNNTLQQIHVHT-------SSDGKSSETEVYYRLQTYKRVLPDVVVAGIPTIN------- 1226
Query 420 ARTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRR 479
R++ N+ +GK E L +G L V+ + T+T++N + E+ DVLGIEA R
Sbjct 1227 -RSVINQESGKIE------LFMEGTGLQAVMNTEGIVGTKTSTNHVMEMKDVLGIEAARY 1279
Query 480 ALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETV 539
+++ E+ ++ G V+ RH+ +L D MT KG ++ ITR G+ +++ L SFE+T
Sbjct 1280 SIISEIGYTMAKHGLTVDPRHIMLLGDVMTCKGEVLGITRFGVAKMKDSVLALASFEKTT 1339
Query 540 EILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLL 577
+ L AA D + G++E I+LG+L P+GT L+
Sbjct 1340 DHLFNAAARFAKDSIEGISECIVLGKLAPIGTNVFQLI 1377
> CE04196
Length=1388
Score = 173 bits (439), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 116/377 (30%), Positives = 191/377 (50%), Gaps = 54/377 (14%)
Query 201 GEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVYLLDEGAYDQERAK 260
GEP+TQMTL TFHFAGV S N+T GVPR+KE+IN KT+ TP +T LLD YD+ A+
Sbjct 1043 GEPSTQMTLKTFHFAGVASMNITQGVPRIKEIINAVKTISTPIITAALLD--PYDESLAR 1100
Query 261 DVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDDEELPQSLGRWLLR 320
V+ +E TTL ++C + +Y PD DY+ L
Sbjct 1101 RVKARIEKTTLGEICDYIEEVYLPD--------------DYF----------------LL 1130
Query 321 VQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDNSDELVLRIRIKKHGQEQDPNS 380
V+L+SK + + L ++ E+I ++ C +I HG+
Sbjct 1131 VKLNSKRI--RLLQLEVCMESISYAIATSKV-CPQMRG--------CKIVAHGKTMMAIR 1179
Query 381 DDQSSEIENRFLQKLMLHCLVGITLRGVSRITKVYMREEARTLYNETTGKFERVNNWVLD 440
+S++ +++ + L + ++G+ + + + + E G F + L
Sbjct 1180 PPSTSKLSKTMTMQILKYSLANVVVKGIPSVNRCVIHAD------EKKGDF-----YSLL 1228
Query 441 TDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRH 500
+G + VL+ VD +T N+ + DVLGIEA R ++ E+ A + G ++ RH
Sbjct 1229 VEGTDFRSVLSSVGVDPRKTNFNNALVVADVLGIEAARSCIINEIIATMDAHGIGLDRRH 1288
Query 501 LAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTEN 560
+ +L D MT +G ++ ITR+G+ +++ L SFE+T++ L EAA F++ D + GV+E
Sbjct 1289 VMLLADVMTYRGEVLGITRNGLVKMKDSVLLLASFEKTMDHLFEAAFFSQRDVIHGVSEC 1348
Query 561 IMLGQLCPLGTGDVDLL 577
I++G +GTG L+
Sbjct 1349 IIMGTPMTVGTGTFKLM 1365
> At5g60040
Length=1328
Score = 168 bits (425), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 119/390 (30%), Positives = 186/390 (47%), Gaps = 59/390 (15%)
Query 188 AGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPSLTVY 247
AG +G I AQSIGEP TQMTL TFHFAGV S N+T GVPR+ E+IN +K + TP ++
Sbjct 980 AGTAIGTIGAQSIGEPGTQMTLKTFHFAGVASMNITQGVPRINEIINASKNISTPVISAE 1039
Query 248 LLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYEFPDD 307
L E + A+ V+ +E TTL +V +V+ + D + +
Sbjct 1040 L--ENPLELTSARWVKGRIEKTTLGQVAESIEVLMTSTSASVRIILDNKII--------- 1088
Query 308 EELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDNSDELVLRI 367
EE S+ W ++ + K P ++ DND I
Sbjct 1089 EEACLSITPWSVKNSI-------LKTPRIKLN-------DND-----------------I 1117
Query 368 RIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCLVGITLRGVSRITKVYMREEARTLYNET 427
R+ G + P D + LH L G+ + +V + E+ +
Sbjct 1118 RVLDTGLDITPVVDKSRAHFN--------LHNLK----NGIKTVERVVVAEDM-----DK 1160
Query 428 TGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRA 487
+ + + W L +G NL V+ P ++ TTSN++ E+ LGIEA R ++ E+
Sbjct 1161 SKQIDGKTKWKLFVEGTNLLAVMGTPGINGRTTTSNNVVEVSKTLGIEAARTTIIDEIGT 1220
Query 488 VISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAV 547
V+ G ++ RH+ +L D MT +G ++ I R GI +++K L + SFE T + L AA
Sbjct 1221 VMGNHGMSIDIRHMMLLADVMTYRGEVLGIQRTGIQKMDKSVLMQASFERTGDHLFSAAA 1280
Query 548 FAETDHLRGVTENIMLGQLCPLGTGDVDLL 577
+ D++ GVTE +++G LGTG + +L
Sbjct 1281 SGKVDNIEGVTECVIMGIPMKLGTGILKVL 1310
> Hs5902062
Length=1391
Score = 167 bits (422), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 138/515 (26%), Positives = 239/515 (46%), Gaps = 86/515 (16%)
Query 81 PINIMRLVEFAKAQFPASVEQLRKQDPVAIAKSVQELLNEKLVVVKQT---CKADTISAE 137
P+ R+++ KA FP P A S EL+ ++K++ C D+ E
Sbjct 918 PLEFKRVLDNIKAVFPC---------PSEPALSKNELILTTESIMKKSEFLCCQDSFLQE 968
Query 138 VQENATIFMKAHLRTVLNSRR--------------LLEREQLGAKAVHWLLGEVERQFHR 183
+++ F+K + +R L + +++ V L ++ R
Sbjct 969 IKK----FIKGVSEKIKKTRDKYGINDNGTTEPRVLYQLDRITPTQVEKFLETCRDKYMR 1024
Query 184 SLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKTVKTPS 243
+ G VGA+ AQSIGEP TQMTL TFHF GV S N+TLGVPR+KE+IN +K + TP
Sbjct 1025 AQMEPGSAVGALCAQSIGEPGTQMTLKTFHFGGVASMNITLGVPRIKEIINASKAISTPI 1084
Query 244 LTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDKQWVSDYYE 303
+T L + D + A+ V+ +E T L ++ + ++ PD +V + +
Sbjct 1085 ITAQLDKDD--DADYARLVKGRIEKTLLGEISEYIEEVFLPDDCFILVKLSLERIR---- 1138
Query 304 FPDDEELPQSLGRWLLRVQLDSKVMIDKKLPMKEIGETIYREFDNDELDCIWTDDNSDEL 363
LLR+++++ ET+ +L D
Sbjct 1139 --------------LLRLEVNA--------------ETVRYSICTSKLRVKPGD------ 1164
Query 364 VLRIRIKKHGQEQDPNSDDQSSEIENRFLQKLMLHCLVGITLRGVSRITKVYMREEARTL 423
+ HG+ + ++S+ ++ + + L + ++G+ +++ +
Sbjct 1165 -----VAVHGEAVVCVTPRENSKSSMYYVLQFLKEDLPKVVVQGIPEVSRAVIH------ 1213
Query 424 YNETTGKFERVNNWVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLR 483
+E +GK + L +G NL V+ V TRTTSN+ E+ LGIEA R ++
Sbjct 1214 IDEQSGK----EKYKLLVEGDNLRAVMATHGVKGTRTTSNNTYEVEKTLGIEAARTTIIN 1269
Query 484 ELRAVISFD-GSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEIL 542
E++ + + G ++ RH+ +L D MT KG ++ ITR G+ ++++ L SFE+T + L
Sbjct 1270 EIQYTMVVNHGMSIDRRHVMLLSDLMTYKGEVLGITRFGLAKMKESVLMLASFEKTADHL 1329
Query 543 MEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDLL 577
+AA F + D + GV+E I++G +GTG LL
Sbjct 1330 FDAAYFGQKDSVCGVSECIIMGIPMNIGTGLFKLL 1364
> 7293125
Length=1259
Score = 107 bits (266), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 52/108 (48%), Positives = 72/108 (66%), Gaps = 2/108 (1%)
Query 177 VERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVA 236
+ +++R++ G VGAIAAQSIGEP TQMTL TFHFAGV S N+T GVPR+ E+IN
Sbjct 886 INDRYNRAVTEPGTAVGAIAAQSIGEPGTQMTLKTFHFAGVASMNITQGVPRIVEIINAT 945
Query 237 KTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDP 284
KT+ TP +T L E + E A+ V+ +E TTL ++ + +V+ P
Sbjct 946 KTISTPIITAEL--ENCHSMEFARQVKARIEKTTLAELSSYVEVVCGP 991
Score = 103 bits (258), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 54/145 (37%), Positives = 84/145 (57%), Gaps = 0/145 (0%)
Query 437 WVLDTDGCNLDEVLTCPRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYV 496
+ L +G L +V+ V RT SN+I EI+ LGIEA R ++ E+ V+ G V
Sbjct 1092 YKLCIEGYGLRDVIATYGVVGKRTRSNNICEIYQTLGIEAARTIIMSEITEVMEGHGMSV 1151
Query 497 NYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRG 556
++RH+ +L MT +G ++ ITRHG+ ++ + SFE+T + L +AA + +TD + G
Sbjct 1152 DWRHIMLLASQMTARGEVLGITRHGLAKMRESVFNLASFEKTADHLFDAAYYGQTDAING 1211
Query 557 VTENIMLGQLCPLGTGDVDLLIDEE 581
V+E I+LG +GTG LL E
Sbjct 1212 VSERIILGMPACIGTGIFKLLQQHE 1236
> ECU04g1400
Length=1395
Score = 97.4 bits (241), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 47/73 (64%), Positives = 59/73 (80%), Gaps = 1/73 (1%)
Query 184 SLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKE-LINVAKTVKTP 242
SLA GE VG IAAQS+GEP+TQMTLNTFH AGVG+KNVTLGVPRL+E LI +K+++TP
Sbjct 957 SLADPGESVGVIAAQSVGEPSTQMTLNTFHLAGVGAKNVTLGVPRLREILITASKSIRTP 1016
Query 243 SLTVYLLDEGAYD 255
+TV + ++D
Sbjct 1017 LITVPIKRRVSFD 1029
Score = 74.7 bits (182), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 40/123 (32%), Positives = 64/123 (52%), Gaps = 1/123 (0%)
Query 455 VDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYL 514
+D SNDI +++ LG+EA R A++ E+ V G ++ RHL ++ D MT+KG
Sbjct 1265 LDIYNAESNDIYDVYLTLGVEAARHAIINEVVKVFDVYGISIDIRHLLLIADYMTRKGSY 1324
Query 515 MSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDV 574
+RHG+ + P+++ SFE AA F D L + ++ +G GTG
Sbjct 1325 SPFSRHGLG-ADDSPIQRISFESCYSNFKTAATFHLEDKLSNPSASLTVGNPVRCGTGCF 1383
Query 575 DLL 577
DL+
Sbjct 1384 DLI 1386
> At3g57660
Length=1670
Score = 95.9 bits (237), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 66/170 (38%), Positives = 93/170 (54%), Gaps = 11/170 (6%)
Query 110 IAKSVQELLNEKLVVVKQTCKADTISAEVQ--ENATIFMKAHLRTVLNSRRLLEREQLGA 167
I K + +N+ +V+ Q C D +S + I +K + + + ER + +
Sbjct 1076 IEKFKELTINQDMVL--QKCSEDMLSGASSYISDLPISLKKGAEKFVEAMPMNER--IAS 1131
Query 168 KAVHW--LLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLG 225
K V LL V+ +F SLA GE VG +AAQS+GEP+TQMTLNTFH AG G NVTLG
Sbjct 1132 KFVRQEELLKLVKSKFFASLAQPGEPVGVLAAQSVGEPSTQMTLNTFHLAGRGEMNVTLG 1191
Query 226 VPRLKE-LINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKV 274
+PRL+E L+ A +KTP +T LL +E A D+ L T+ +
Sbjct 1192 IPRLQEILMTAAANIKTPIMTCPLLK--GKTKEDANDITDRLRKITVADI 1239
Score = 70.1 bits (170), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 62/124 (50%), Gaps = 0/124 (0%)
Query 454 RVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGY 513
++D SN I ++ ++ G+EA R ++RE+ V G V+ RHL ++ D MT G
Sbjct 1542 KLDVRYLYSNSIHDMLNIFGVEAARETIIREINHVFKSYGISVSIRHLNLIADYMTFSGG 1601
Query 514 LMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPLGTGD 573
++R G P + +FE + +++AA + E D L + I LG GTG
Sbjct 1602 YRPMSRMGGIAESTSPFCRMTFETATKFIVQAATYGEKDTLETPSARICLGLPALSGTGC 1661
Query 574 VDLL 577
DL+
Sbjct 1662 FDLM 1665
> SPBC4C3.05c
Length=1689
Score = 94.7 bits (234), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 43/75 (57%), Positives = 60/75 (80%), Gaps = 1/75 (1%)
Query 177 VERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVA 236
++ ++ +SL GE VG +A+QSIGEP+TQMTLNTFHFAG G+KNVTLG+PRL+E+I A
Sbjct 1179 MQLRYQQSLVDPGESVGVLASQSIGEPSTQMTLNTFHFAGFGAKNVTLGIPRLREIIMTA 1238
Query 237 K-TVKTPSLTVYLLD 250
++TP++T+ L D
Sbjct 1239 SANIQTPTMTLRLND 1253
Score = 78.6 bits (192), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 69/138 (50%), Gaps = 2/138 (1%)
Query 441 TDGCNLDEVLTC-PRVDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYR 499
T+G NL + + +NDI I + G+EA R A++ E+ +V G V+ R
Sbjct 1544 TEGVNLKAIWEFYNEISMNDIYTNDIAAILRIYGVEAARNAIVHEVSSVFGVYGIAVDPR 1603
Query 500 HLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTE 559
HL+++ D MT +G + R GI P K SFE T L EAA+ + D L +
Sbjct 1604 HLSLIADYMTFEGGYKAFNRMGI-EYNTSPFAKMSFETTCHFLTEAALRGDVDDLSNPSS 1662
Query 560 NIMLGQLCPLGTGDVDLL 577
+++G++ GTG D+
Sbjct 1663 RLVVGRVGNFGTGSFDIF 1680
> Hs7661686
Length=1717
Score = 94.0 bits (232), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 54/129 (41%), Positives = 81/129 (62%), Gaps = 8/129 (6%)
Query 177 VERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVA 236
++ ++ RSL GE VG +AAQSIGEP+TQMTLNTFHFAG G NVTLG+PRL+E++ VA
Sbjct 1197 LQLKWQRSLCEPGEAVGLLAAQSIGEPSTQMTLNTFHFAGRGEMNVTLGIPRLREILMVA 1256
Query 237 K-TVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQVIYDPDPTKTIVPQDK 295
+KTP ++V +L+ +R K ++ L +VC + +V+ D ++ ++K
Sbjct 1257 SANIKTPMMSVPVLNTKK-ALKRVKSLKKQ-----LTRVC-LGEVLQKIDVQESFCMEEK 1309
Query 296 QWVSDYYEF 304
Q Y+
Sbjct 1310 QNKFQVYQL 1318
Score = 82.0 bits (201), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 54/157 (34%), Positives = 85/157 (54%), Gaps = 4/157 (2%)
Query 421 RTLYNETTGKFERVNNWVLDTDGCNLDEVLTCPRV-DSTRTTSNDITEIFDVLGIEAVRR 479
R L NETT + VL+T+G NL E+ V D R SNDI I + GIEA+R
Sbjct 1559 RCLLNETTNN-KNEKELVLNTEGINLPELFKYAEVLDLRRLYSNDIHAIANTYGIEALR- 1616
Query 480 ALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETV 539
+ +E++ V + G V+ RHL+++ D M +G + R GI R PL++ +FE +
Sbjct 1617 VIEKEIKDVFAVYGIAVDPRHLSLVADYMCFEGVYKPLNRFGI-RSNSSPLQQMTFETSF 1675
Query 540 EILMEAAVFAETDHLRGVTENIMLGQLCPLGTGDVDL 576
+ L +A + D LR + +++G++ GTG +L
Sbjct 1676 QFLKQATMLGSHDELRSPSACLVVGKVVRGGTGLFEL 1712
> CE22157
Length=1737
Score = 92.8 bits (229), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 63/184 (34%), Positives = 102/184 (55%), Gaps = 23/184 (12%)
Query 95 FPASVEQLRKQDPVAIAKSVQELLNEKLVVVKQTCKADTISAEVQENATIFMKAHLRTVL 154
F S+E+ R+Q I K E ++E+ TCK + ++ + F ++ V
Sbjct 1152 FELSLEE-REQYARGIPKKCPEAVDERF---NPTCKLGALPEKMLDEIEGFCTRRVKKVD 1207
Query 155 NS---RRLLEREQLGAKAVHWLLGEVERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNT 211
+ + +L+R ++W + RSLA GE VG +AAQSIGEP+TQMTLNT
Sbjct 1208 DEEPPKEVLKR------TLYW-------KGMRSLADPGENVGLLAAQSIGEPSTQMTLNT 1254
Query 212 FHFAGVGSKNVTLGVPRLKE-LINVAKTVKTPSLTVYLLDEGAYDQERAKDVQTHLEHTT 270
FHFAG G NVTLG+PRL+E L+ +K++ TPS ++ ++ ++R ++ L+
Sbjct 1255 FHFAGRGEMNVTLGIPRLREILMTASKSIATPSASIAVI--AGTSRDRIDSIKRELDRVY 1312
Query 271 LEKV 274
L+++
Sbjct 1313 LKQL 1316
Score = 76.6 bits (187), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/143 (33%), Positives = 71/143 (49%), Gaps = 2/143 (1%)
Query 438 VLDTDGCNLDEVLTCPRV-DSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYV 496
+L T G NL V D SND+ I + G+EA +A+ E+ V + G V
Sbjct 1572 ILQTQGVNLAAFFKHADVLDVNSVYSNDLNLILENYGVEACSKAITTEMNNVFAVYGIEV 1631
Query 497 NYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRG 556
+ RHL++ D MT G + R G PL+K +FE T+ L EA + E D++
Sbjct 1632 SKRHLSLTADYMTFTGQIQPFNR-GAMSSSSSPLQKMTFETTMAFLREALLHGEEDNVNS 1690
Query 557 VTENIMLGQLCPLGTGDVDLLID 579
+ +++G L GTG DLL+D
Sbjct 1691 PSARLVMGALPRGGTGSFDLLLD 1713
> YOR341w
Length=1664
Score = 92.0 bits (227), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 63/81 (77%), Gaps = 1/81 (1%)
Query 177 VERQFHRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVA 236
++ ++ RSL + GE VG IA+QS+GEP+TQMTLNTFHFAG G+ NVTLG+PRL+E++ A
Sbjct 1170 MQLKYMRSLINPGEAVGIIASQSVGEPSTQMTLNTFHFAGHGAANVTLGIPRLREIVMTA 1229
Query 237 K-TVKTPSLTVYLLDEGAYDQ 256
+KTP +T+ + ++ + +Q
Sbjct 1230 SAAIKTPQMTLPIWNDVSDEQ 1250
Score = 74.7 bits (182), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 43/141 (30%), Positives = 72/141 (51%), Gaps = 2/141 (1%)
Query 438 VLDTDGCNLDEVLTCPR-VDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYV 496
VL T+G N + +D TSND+ + G+EA R ++ E+ V S V
Sbjct 1518 VLVTEGVNFQAMWDQEAFIDVDGITSNDVAAVLKTYGVEAARNTIVNEINNVFSRYAISV 1577
Query 497 NYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRG 556
++RHL ++ D MT++G ++ R G+ K S+E T + L +A + E + L
Sbjct 1578 SFRHLDLIADMMTRQGTYLAFNRQGM-ETSTSSFMKMSYETTCQFLTKAVLDNEREQLDS 1636
Query 557 VTENIMLGQLCPLGTGDVDLL 577
+ I++G+L +GTG D+L
Sbjct 1637 PSARIVVGKLNNVGTGSFDVL 1657
> 7303170
Length=1642
Score = 92.0 bits (227), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 49/99 (49%), Positives = 67/99 (67%), Gaps = 4/99 (4%)
Query 183 RSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGVGSKNVTLGVPRLKELINVAKT-VKT 241
+SLA GE VG IAAQSIGEP+TQMTLNTFHFAG G NVTLG+PRL+E++ +A + +KT
Sbjct 1169 KSLAAPGEPVGLIAAQSIGEPSTQMTLNTFHFAGRGEMNVTLGIPRLREILMLASSNIKT 1228
Query 242 PSLTVYLLDEGAYDQERAKDVQTHLEHTTLEKVCTMAQV 280
PS+ + + Q +A+ ++ +L TL + V
Sbjct 1229 PSMDIPI---KPGQQHQAEKLRINLNSVTLANLLEYVHV 1264
Score = 81.6 bits (200), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 48/144 (33%), Positives = 76/144 (52%), Gaps = 2/144 (1%)
Query 435 NNWVLDTDGCNLDEVLTCPRV-DSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDG 493
++ +L TDG N+ E+ ++ D R SNDI I GIEA + +++E+ V G
Sbjct 1496 DDQLLKTDGINIGEMFQHNKILDLNRLYSNDIHAIARTYGIEAASQVIVKEVSNVFKVYG 1555
Query 494 SYVNYRHLAILCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDH 553
V+ RHL+++ D MT G ++R G+ PL++ SFE +++ L AA F D
Sbjct 1556 ITVDRRHLSLIADYMTFDGTFQPLSRKGMEH-SSSPLQQMSFESSLQFLKSAAGFGRADE 1614
Query 554 LRGVTENIMLGQLCPLGTGDVDLL 577
L + +M+G GTG +LL
Sbjct 1615 LSSPSSRLMVGLPVRNGTGAFELL 1638
> AtCh013
Length=1376
Score = 50.8 bits (120), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 182 HRSLAHAGECVGAIAAQSIGEPATQMTLNTFHFAGV 217
H L GE VG IA QSIGEP TQ+TL TFH GV
Sbjct 310 HGDLVELGEAVGIIAGQSIGEPGTQLTLRTFHTGGV 345
Score = 48.5 bits (114), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 41/69 (59%), Gaps = 2/69 (2%)
Query 504 LCDTMTQKGYLMSITRHGINRVEKGPLKKCSFEETVEILMEAAVFAETDHLRGVTENIML 563
L + + + L+ ITR +N + + + SF+ET +L +AA+ D L+G+ EN++L
Sbjct 1256 LEEAICYRAVLLGITRASLN--TQSFISEASFQETARVLAKAALRGRIDWLKGLKENVVL 1313
Query 564 GQLCPLGTG 572
G + P GTG
Sbjct 1314 GGVIPAGTG 1322
> At1g63020_1
Length=1316
Score = 48.1 bits (113), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/124 (22%), Positives = 58/124 (46%), Gaps = 5/124 (4%)
Query 455 VDSTRTTSNDITEIFDVLGIEAVRRALLRELRAVISFDGSYVNYRHLAILCDTMTQKGYL 514
+D R+ ++I + V GI+A R + L + +S G + HL ++ D+++ G
Sbjct 1116 IDWGRSHPDNIRQCCSVYGIDAGRSIFVANLESAVSDTGKEILREHLLLVADSLSVTGEF 1175
Query 515 MSITRHGINRVEK-----GPLKKCSFEETVEILMEAAVFAETDHLRGVTENIMLGQLCPL 569
+++ G ++ + P + F + ++AA D L+G + + G++
Sbjct 1176 VALNAKGWSKQRQVESTPAPFTQACFSSPSQCFLKAAKEGVRDDLQGSIDALAWGKVPGF 1235
Query 570 GTGD 573
GTGD
Sbjct 1236 GTGD 1239
> CE24451
Length=641
Score = 34.7 bits (78), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 64/151 (42%), Gaps = 16/151 (10%)
Query 76 VKQHIPINIMRLVEFAKAQFPASVEQLRK---QD-----PVAIAKSVQELLNEKLVVV-- 125
V Q +P R A FP ++ L K +D V S E + ++L+ V
Sbjct 303 VGQGMPPKTARTTAMFSATFPKEIQVLAKDFLKDNYIFLAVGRVGSTSENIEQRLLWVNE 362
Query 126 --KQTCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQLGAKAVHWLLGEVERQFHR 183
K++ + + E EN + R L R+Q+ + ++H L ++ER+ +
Sbjct 363 MEKRSNLMEILMNEHSENLVLVFVETKRGANELAYFLNRQQIRSVSIHGDLKQIERERNL 422
Query 184 SLAHAGEC----VGAIAAQSIGEPATQMTLN 210
L +G+C A+AA+ + P + +N
Sbjct 423 ELFRSGQCPILVATAVAARGLDIPNVRHVIN 453
> CE28369
Length=644
Score = 34.7 bits (78), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 64/151 (42%), Gaps = 16/151 (10%)
Query 76 VKQHIPINIMRLVEFAKAQFPASVEQLRK---QD-----PVAIAKSVQELLNEKLVVV-- 125
V Q +P R A FP ++ L K +D V S E + ++L+ V
Sbjct 306 VGQGMPPKTARTTAMFSATFPKEIQVLAKDFLKDNYIFLAVGRVGSTSENIEQRLLWVNE 365
Query 126 --KQTCKADTISAEVQENATIFMKAHLRTVLNSRRLLEREQLGAKAVHWLLGEVERQFHR 183
K++ + + E EN + R L R+Q+ + ++H L ++ER+ +
Sbjct 366 MEKRSNLMEILMNEHSENLVLVFVETKRGANELAYFLNRQQIRSVSIHGDLKQIERERNL 425
Query 184 SLAHAGEC----VGAIAAQSIGEPATQMTLN 210
L +G+C A+AA+ + P + +N
Sbjct 426 ELFRSGQCPILVATAVAARGLDIPNVRHVIN 456
> 7295084
Length=568
Score = 32.0 bits (71), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 32/77 (41%), Gaps = 2/77 (2%)
Query 403 ITLRGVSRITKVYMREEARTLYNETTGK--FERVNNWVLDTDGCNLDEVLTCPRVDSTRT 460
IT S +V E RTL +G+ F N ++LD GC L V T + +
Sbjct 255 ITKLDASSSARVAKSESPRTLQRTLSGRTLFVSGNKFILDPSGCRLTRVSTSSTGATQSS 314
Query 461 TSNDITEIFDVLGIEAV 477
+ I D+ G+ V
Sbjct 315 VNRSILRRIDIGGLTYV 331
> At1g68920
Length=486
Score = 30.8 bits (68), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 31/63 (49%), Gaps = 3/63 (4%)
Query 590 VEPQQDLALEGLASPGGADGMSPDGSGTPSPHNVTSVFSPMP---FSTVYSSFAQSPSNP 646
V PQ D LEGL + + S TP P N++ + P+P SS ++ ++P
Sbjct 373 VNPQMDFNLEGLLAKDALQLRAGSSSTTPFPPNMSMAYPPLPHGFMQQTLSSIGRTITSP 432
Query 647 LSP 649
LSP
Sbjct 433 LSP 435
Lambda K H
0.317 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18603415754
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40