bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0999_orf3
Length=366
Score E
Sequences producing significant alignments: (Bits) Value
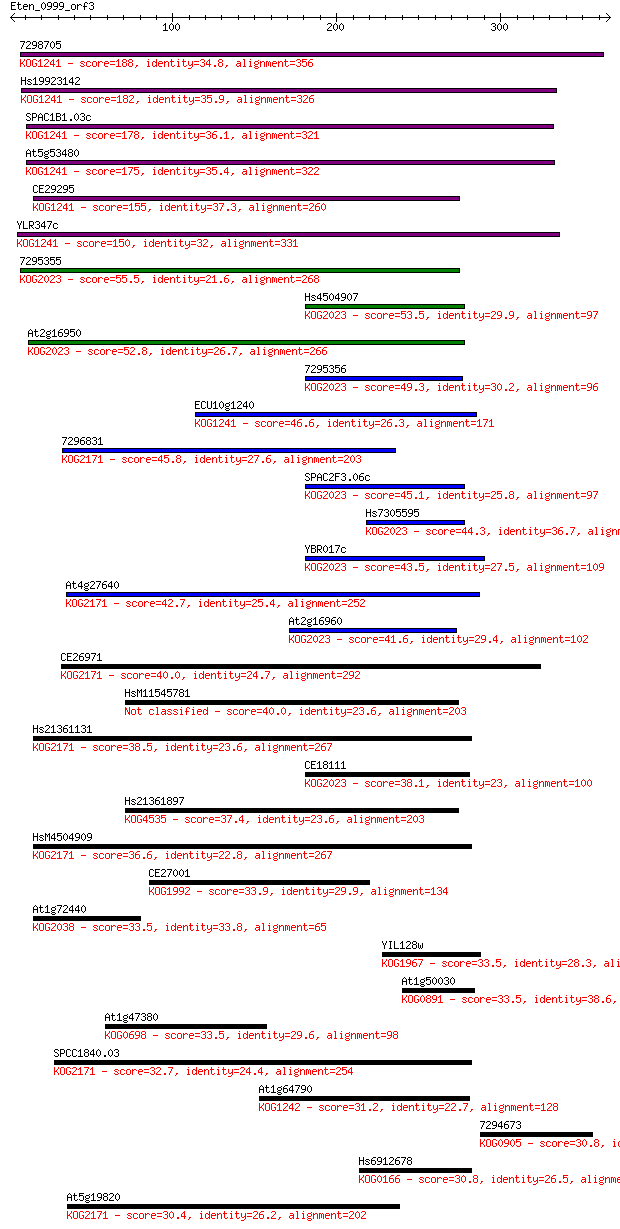
7298705 188 2e-47
Hs19923142 182 9e-46
SPAC1B1.03c 178 1e-44
At5g53480 175 1e-43
CE29295 155 1e-37
YLR347c 150 5e-36
7295355 55.5 2e-07
Hs4504907 53.5 8e-07
At2g16950 52.8 1e-06
7295356 49.3 1e-05
ECU10g1240 46.6 9e-05
7296831 45.8 2e-04
SPAC2F3.06c 45.1 2e-04
Hs7305595 44.3 4e-04
YBR017c 43.5 8e-04
At4g27640 42.7 0.001
At2g16960 41.6 0.003
CE26971 40.0 0.008
HsM11545781 40.0 0.008
Hs21361131 38.5 0.024
CE18111 38.1 0.033
Hs21361897 37.4 0.058
HsM4504909 36.6 0.082
CE27001 33.9 0.64
At1g72440 33.5 0.65
YIL128w 33.5 0.75
At1g50030 33.5 0.77
At1g47380 33.5 0.83
SPCC1840.03 32.7 1.4
At1g64790 31.2 3.8
7294673 30.8 4.6
Hs6912678 30.8 4.8
At5g19820 30.4 6.0
> 7298705
Length=884
Score = 188 bits (477), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 124/372 (33%), Positives = 200/372 (53%), Gaps = 44/372 (11%)
Query 7 EDPSEALSQQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYI 64
+D + + + N VLTA+ HG++ +P +++AA AL+++L F K NF K ER++I
Sbjct 165 QDIRFGVMENQSNDVLTAIIHGMRKVEPSNHVRLAATTALHNSLEFTKSNFEKDMERNFI 224
Query 65 ISSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVI 123
+ V EA + S + VAAL+C+V+I YY ++ YM A+ +T A+KS + AV +
Sbjct 225 MEVVCEATQCQD-SQICVAALQCLVKIMTLYYQYMEPYMAQALFPITLAAMKSDNDAVAL 283
Query 124 AALEVWNALAEEEL--------AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDE 175
+E W+ + +EE+ A +G + + + + A FL P+L+E+L +DE
Sbjct 284 QGIEFWSNVCDEEIDLAIESQEATDQGRAPQRVSKHYARGALQFLTPVLVEKL--TKQDE 341
Query 176 -DDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVME 234
DD+D+W+PA AA++CL + A +D I+P V+ F++ N S WR R+A+V+ G V+
Sbjct 342 CDDEDTWSPAKAASVCLMVLATCCEDEIVPHVLPFIKENIESPNWRFRDAAVMTFGSVLN 401
Query 235 GPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVS 294
G + P V ++ P L+ + D+SV VRD+ AWT G+ + P A ++
Sbjct 402 GLETNTLKPLVEQAMPTLIR-LMYDSSVIVRDTIAWTFGRICD---------IIPEAAIN 451
Query 295 STTSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDINTPEGRAAYQGEAATQQQD 354
T LL V+ L +PRVA ++CW L+D AA+ EAA
Sbjct 452 ETYLQ-----TLLECFVKSLKSEPRVAANVCWAFIGLSD--------AAW--EAAVTNDG 496
Query 355 QTP----LDPIF 362
+TP L P F
Sbjct 497 ETPETYALSPYF 508
> Hs19923142
Length=876
Score = 182 bits (463), Expect = 9e-46, Method: Compositional matrix adjust.
Identities = 117/337 (34%), Positives = 186/337 (55%), Gaps = 30/337 (8%)
Query 8 DPSEALSQQRCNVVLTAVAHGLKDPDP--QLKVAALRALYHALIFAKENFAKTEERDYII 65
DP + Q + N +LTA+ G++ +P +K+AA AL ++L F K NF K ER +I+
Sbjct 162 DPEQL--QDKSNEILTAIIQGMRKEEPSNNVKLAATNALLNSLEFTKANFDKESERHFIM 219
Query 66 SSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIA 124
V EA + + V+VAAL+ +V+I YY ++ YM PA+ ++T A+KS V +
Sbjct 220 QVVCEATQCPD-TRVRVAALQNLVKIMSLYYQYMETYMGPALFAITIEAMKSDIDEVALQ 278
Query 125 ALEVWNALAEEEL--------AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDED 176
+E W+ + +EE+ A +G T K A +L+P+L Q L ++ D
Sbjct 279 GIEFWSNVCDEEMDLAIEASEAAEQGRPPEHTSKFYAKGALQYLVPIL-TQTLTKQDEND 337
Query 177 DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGP 236
DDD W P AA +CL L A +D I+P V+ F++ + + +WR R+A+V+A GC++EGP
Sbjct 338 DDDDWNPCKAAGVCLMLLATCCEDDIVPHVLPFIKEHIKNPDWRYRDAAVMAFGCILEGP 397
Query 237 SAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSST 296
+ P V ++ P L+E + D SV VRD+AAWT+G+ + L P A ++
Sbjct 398 EPSQLKPLVIQAMPTLIE-LMKDPSVVVRDTAAWTVGRICE---------LLPEAAINDV 447
Query 297 TSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELAD 333
LL ++E L+ +PRVA ++CW LA+
Sbjct 448 YL-----APLLQCLIEGLSAEPRVASNVCWAFSSLAE 479
> SPAC1B1.03c
Length=863
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 116/333 (34%), Positives = 182/333 (54%), Gaps = 31/333 (9%)
Query 11 EALSQQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSV 68
E LS Q N +LTAV G + +PD +++AAL ALY +L F +ENF ER+YI+ V
Sbjct 164 EVLSAQ-SNAILTAVVAGARKEEPDAAVRLAALGALYDSLEFVRENFNNEYERNYIMQVV 222
Query 69 LEAGNAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIAALE 127
EA + S +Q AA C+V+I YY + YM A+ ++T + +++ V + A+E
Sbjct 223 CEATQSPEAS-IQTAAFGCLVKIMHLYYDTMPFYMEKALFALTTQGMYNTNEQVALQAVE 281
Query 128 VWNALAEEELAIIEGESTRWTLLN---------IMKEATPFLLPLLLEQLLNAAEDEDDD 178
W+ + EEE+ + E LN + A +LP+LL+ L N ED D+D
Sbjct 282 FWSTVCEEEIEV-NLEIQEAQDLNEVPARQNHGFARAAAADILPVLLKLLCNQDEDADED 340
Query 179 DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSA 238
D W +MAAA CL L AQVV D I+ V+ FV+ N + +W +REA+V+A G V+EGP+
Sbjct 341 D-WNISMAAATCLQLFAQVVGDLIVNPVLAFVEQNIQNPDWHQREAAVMAFGSVLEGPNV 399
Query 239 EAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTS 298
+ P V ++ P+L+ + D + V+D+ AW LG+ + + V+ +
Sbjct 400 AMLTPLVNQALPVLINMMV-DPVIFVKDTTAWALGQIS--------------SFVADAIN 444
Query 299 PNNNGGALLLAIVERLTDQPRVAVHICWMLHEL 331
P + ++ A+++ LTD PR+ + CW L
Sbjct 445 PEIHLSPMVSALLQGLTDNPRIVANCCWAFMNL 477
> At5g53480
Length=870
Score = 175 bits (444), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 114/334 (34%), Positives = 180/334 (53%), Gaps = 25/334 (7%)
Query 11 EALSQQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSV 68
+ + Q+ N +LTAV G+ + + +++AA RALY AL FA+ NF ERDYI+ V
Sbjct 165 DVVEQEHVNKILTAVVQGMNAAEGNTDVRLAATRALYMALGFAQANFNNDMERDYIMRVV 224
Query 69 LEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEV 128
EA + ++ AA EC+V I+ YY L YM + ++T A++ D +V + A+E
Sbjct 225 CEA-TLSPEVKIRQAAFECLVSIASTYYEKLAHYMQDIFNITAKAVREDDESVALQAIEF 283
Query 129 WNALAEEELAIIEGESTRWT------LLNIMKEATPFLLPLLLEQLLNAAEDED-DDDSW 181
W+++ +EE+ I+E + K+A P L+PLLLE LL ED+D D+ +W
Sbjct 284 WSSICDEEIDILEEYGGEFAGDSDVPCFYFTKQALPGLVPLLLETLLKQEEDQDLDEGAW 343
Query 182 TPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAM 241
AMA CLGL A+ V D I+P V+ F++ +WR REA+ A G ++EGPSA+ +
Sbjct 344 NIAMAGGTCLGLVARAVGDDIVPHVMPFIEEKISKPDWREREAATYAFGSILEGPSADKL 403
Query 242 APYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSP-- 299
V + ++ A+ +D S V+D+ AWTLG+ +F S+ +P
Sbjct 404 MAIVNAALTFMLNALTNDPSNHVKDTTAWTLGR------------IFEFLHGSTIETPII 451
Query 300 -NNNGGALLLAIVERLTDQPRVAVHICWMLHELA 332
N ++ +++ + D P VA C L+ LA
Sbjct 452 NQANCQQIITVLIQSMNDAPNVAEKACGALYFLA 485
> CE29295
Length=883
Score = 155 bits (392), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 97/272 (35%), Positives = 163/272 (59%), Gaps = 16/272 (5%)
Query 15 QQRCNVVLTAVAHGLK--DPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAG 72
+ + N VLTA+ HG++ + ++ AA AL ++L F NF+ ER+ I+ V E+
Sbjct 173 ETKANDVLTAIIHGMRPEESSANVRFAATNALLNSLEFTNTNFSNEAERNIIMQVVCEST 232
Query 73 NAANPSAVQVAALECIVQISQEYYSFLGAYM-PAVGSVTWNALKSSDSAVVIAALEVWNA 131
++++ V+VAAL+C+V+I Q YY + +YM A+ +T +A+KS + V + +E W+
Sbjct 233 SSSD-QRVKVAALQCLVRIMQLYYEHMLSYMGSALFQITLSAMKSQEPEVAMQGMEFWST 291
Query 132 LAEEELAII---EGESTR------WTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWT 182
+AEEE + E E R L M++A + P+LLE + + + +DDD WT
Sbjct 292 VAEEEFDLYMTYEDEVERGAPNPKCASLRFMEQAASHVCPVLLEAMAHHDDGDDDD-DWT 350
Query 183 PAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMA 242
PA AA +CL L AQ V+D I+ VI F + +F + +W+ +EA+++A G +++GP + +
Sbjct 351 PAKAAGVCLMLAAQCVRDDIVNHVIPFFK-HFQNPDWKYKEAAIMAFGSILDGPDPKKLL 409
Query 243 PYVARSFPILVEAVASDASVAVRDSAAWTLGK 274
P + P +V A+ D +V VRD+AAW+LG+
Sbjct 410 PMAQEALPAIVAAMC-DKNVNVRDTAAWSLGR 440
> YLR347c
Length=861
Score = 150 bits (378), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 106/343 (30%), Positives = 170/343 (49%), Gaps = 34/343 (9%)
Query 5 EGEDPSEALSQQRCNVVLTAVAHGLKDPDPQ--LKVAALRALYHALIFAKENFAKTEERD 62
E DP N +L A+ G + + +++AAL AL +LIF K N + ER+
Sbjct 164 ESADPQSQALVSSSNNILIAIVQGAQSTETSKAVRLAALNALADSLIFIKNNMEREGERN 223
Query 63 YIISSVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMP-AVGSVTWNALKSSDSAV 121
Y++ V EA A + VQ AA C+ +I YY+F+ YM A+ ++T +KS + V
Sbjct 224 YLMQVVCEATQAEDIE-VQAAAFGCLCKIMSLYYTFMKPYMEQALYALTIATMKSPNDKV 282
Query 122 VIAALEVWNALAEEELAIIEG---------ESTRWTLLNIMKEATPFLLPLLLEQLLNAA 172
+E W+ + EEE+ I +S + L +I K+ P LL LL Q
Sbjct 283 ASMTVEFWSTICEEEIDIAYELAQFPQSPLQSYNFALSSI-KDVVPNLLNLLTRQ----N 337
Query 173 EDEDDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCV 232
ED +DDD W +M+A CL L AQ + IL V+ FV+ N + WR REA+V+A G +
Sbjct 338 EDPEDDD-WNVSMSAGACLQLFAQNCGNHILEPVLEFVEQNITADNWRNREAAVMAFGSI 396
Query 233 MEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPAD 292
M+GP YV ++ P ++ + +D S+ V+++ AW +G+ A
Sbjct 397 MDGPDKVQRTYYVHQALPSILN-LMNDQSLQVKETTAWCIGRIAD--------------S 441
Query 293 VSSTTSPNNNGGALLLAIVERLTDQPRVAVHICWMLHELADDI 335
V+ + P + ++ A + L D P+VA + W + L + +
Sbjct 442 VAESIDPQQHLPGVVQACLIGLQDHPKVATNCSWTIINLVEQL 484
> 7295355
Length=884
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 58/300 (19%), Positives = 121/300 (40%), Gaps = 37/300 (12%)
Query 7 EDPSEALSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIIS 66
ED + L N ++ K P+++ A+ + +I + A D +I
Sbjct 163 EDSAGILENMPLNTMIPKFLEYFKHSSPKIRSHAIACINQFIINRSQ--ALMLNIDSLIQ 220
Query 67 SVLEAGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAAL 126
++L+ + +P AV++ +V + + +M + + + +D V + A
Sbjct 221 NLLDVPSDNDP-AVRMNVCHALVGLVGVRIDLMMPHMSQIIELILLRSQDADENVALQAT 279
Query 127 EVWNALAEE------------ELAIIEGESTRWTLLNIM----------------KEATP 158
E W +L ++ +L + T++T +I+ ++ +P
Sbjct 280 EFWLSLGKQRNCRDILSPILSQLVPVLVSRTQYTETDIILLKGDVGEDDKEPDRQQDISP 339
Query 159 FLLPLLLEQLLNAAEDEDDDDSWTPAMAAAIC----LGLCAQVVKDAILPLVINFVQLNF 214
+ + N ++ DDD W A+ C L + +++ D LPL++ ++
Sbjct 340 RFHMSRVHGISNELDENSDDDMWDSALNLRKCSACALDIISKIFGDVCLPLMLPILKEAL 399
Query 215 GSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGK 274
EW +E+ V+ALG + EG + + P++ P L+ + SD VR WT +
Sbjct 400 FHQEWVIKESGVMALGAIAEG-CMQGLIPHLPELIPYLITCL-SDKKPLVRSITCWTFMR 457
> Hs4504907
Length=890
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 47/97 (48%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V +D +LP ++ ++ EW +E+ +L LG + EG +
Sbjct 373 WNLRKCSAAALDVLANVYRDELLPHILPLLKELLFHHEWVVKESGILVLGAIAEG-CMQG 431
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M PY+ P L++ + SD VR WTL + A
Sbjct 432 MIPYLPELIPHLIQCL-SDKKALVRSITCWTLSRYAH 467
> At2g16950
Length=827
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 71/315 (22%), Positives = 117/315 (37%), Gaps = 70/315 (22%)
Query 12 ALSQQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEA 71
L+++ N+ L + + P L+ AL ++ +I + ++ VL
Sbjct 173 GLAERPINIFLPRLLQFFQSPHASLRKLALGSVNQYIIIMPAALYNSLDKYLQGLFVL-- 230
Query 72 GNAANPSAVQVAALEC--IVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVW 129
AN +V L C V +++ S + ++ V + D V + A E W
Sbjct 231 ---ANDPVPEVRKLVCAAFVHLTEVLPSSIEPHLRNVMEYMLQVNRDPDEEVSLEACEFW 287
Query 130 NALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQ---------LLNAAEDEDDDDS 180
+A + +L +KE P L+P+LLE LL+A EDE D
Sbjct 288 SAYCDAQLPP-----------ENLKEFLPRLIPVLLENMAYADDDESLLDAEEDESQPDR 336
Query 181 -----------------------------WTPAMAAAICLGLCAQVVKDAILPLVINFVQ 211
W +A + + + V D ILP ++ +Q
Sbjct 337 DQDLKPRFHTSRLHGSEDFDDDDDDSFNVWNLRKCSAAAIDVLSNVFGDEILPALMPLIQ 396
Query 212 LNF---GSTEWRRREASVLALGCVMEG------PSAEAMAPYVARSFPILVEAVASDASV 262
N G W++REA+VLALG + EG P + VA P+L D
Sbjct 397 KNLSASGDEAWKQREAAVLALGAIAEGCMNGLYPHLSEASLIVAFLLPLL-----DDKFP 451
Query 263 AVRDSAAWTLGKTAQ 277
+R + WTL + +
Sbjct 452 LIRSISCWTLSRFGK 466
> 7295356
Length=893
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A V ++ LP+V+ ++ EW +E+ VLALG + EG +
Sbjct 377 WNLRKCSAAALDVLANVFREDCLPVVLPILKETLFHQEWVIKESGVLALGAIAEG-CMQG 435
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTA 276
M ++ P L+ + SD VR WTL + A
Sbjct 436 MIQHLPELIPYLISCL-SDKKALVRSITCWTLSRYA 470
> ECU10g1240
Length=854
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 45/180 (25%), Positives = 77/180 (42%), Gaps = 30/180 (16%)
Query 114 LKSSDSAVVIAALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAE 173
KS + I A+E W AE+ +GE ++ + P +LP +L L
Sbjct 296 FKSQHDEIKIQAIEYWCIFAEKN----DGE--------MVDKYLPVVLPEILSLLEKGPN 343
Query 174 DEDDDDSWTPAMAAAICLGLCAQVVKDAIL--PLVINFVQLNFGSTEWRRREASVLALGC 231
D W+P AA+ CL + ++ KD ++ +V F++ + S + +ALG
Sbjct 344 YYGD--VWSPHKAASSCLEMYTELKKDKMMRNRMVWGFIETSLRSESRANIDIGAVALGS 401
Query 232 VMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ-------QHAPIVL 284
VM + + V P LV+ + + S +DS W L + A+ H P++L
Sbjct 402 VMHERCEDCLTKIV----PDLVKGIEFEES---KDSCLWALSRAAECNFYALADHLPMIL 454
> 7296831
Length=1105
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 56/235 (23%), Positives = 100/235 (42%), Gaps = 60/235 (25%)
Query 33 DPQLKVAALRA-----LYHALIFAKEN-------FAKTEERD-YIISSVLEAGNAANPSA 79
DP+++V A+RA LYH KEN FA R +I +EA +
Sbjct 183 DPEVRVQAVRAVGAFILYHD----KENETAIHKHFADMLPRMIHITGETIEAQDDQ---- 234
Query 80 VQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSD--SAVVIAALEVWNALAEEEL 137
+ L+ ++++++ FL + + V S D + LEV +LAE
Sbjct 235 ---SLLKLLIEMTENCPKFLRPQLEFIFEVCMKVFSSQDFEDSWRHLVLEVMVSLAENAP 291
Query 138 AIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAA--------- 188
++I + ++ + L+PL+L + D DDD++W+ A
Sbjct 292 SMIRKRADKYIVA---------LIPLILHMM----TDLDDDENWSTADVVDDDDHSDNNV 338
Query 189 --------ICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEG 235
+ GL ++V LPLV+N + + G +W+ R A+++A+ + EG
Sbjct 339 IAESSLDRLACGLGGKIV----LPLVMNALPVMLGHADWKHRFAALMAISAIGEG 389
> SPAC2F3.06c
Length=910
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 49/97 (50%), Gaps = 2/97 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + + K +L +++ ++ + S +W+ +EA VLA+G + EG +
Sbjct 390 WNLRKCSAAALDVLSSFWKQRLLEIILPHLKQSLTSEDWKVQEAGVLAVGAIAEG-CMDG 448
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
M Y+ +P + + S + VR WTLG+ ++
Sbjct 449 MVQYLPELYPYFLSLLDSKKPL-VRTITCWTLGRYSK 484
> Hs7305595
Length=887
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 218 EWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQ 277
EW +E+ +L LG + EG + M PY+ P L++ + SD VR A WTL + A
Sbjct 407 EWVVKESGILVLGAIAEG-CMQGMVPYLPELIPHLIQCL-SDKKALVRSIACWTLSRYAH 464
> YBR017c
Length=918
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 51/113 (45%), Gaps = 6/113 (5%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + ++ ++ + F++ + GS W REA++LALG + EG +
Sbjct 397 WNLRKCSAATLDVMTNILPHQVMDIAFPFLREHLGSDRWFIREATILALGAMAEG-GMKY 455
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTA----QQHAPIVLRHLFP 289
+ P LVE + +D VR WTL + + Q H ++ L P
Sbjct 456 FNDGLPALIPFLVEQL-NDKWAPVRKMTCWTLSRFSPWILQDHTEFLIPVLEP 507
> At4g27640
Length=651
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 64/258 (24%), Positives = 119/258 (46%), Gaps = 22/258 (8%)
Query 35 QLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN---AANPSAVQVAALECIVQI 91
+++VAAL+A+ L F + + RD+I S+L+ A+ V + A E ++
Sbjct 175 RVRVAALKAVGSFLEFTNDGDEVVKFRDFI-PSILDVSRKCIASGEEDVAILAFEIFDEL 233
Query 92 SQEYYSFLGAYMPAV--GSVTWNALKSSDSAVVIAALEVWNALAEEELAIIEGESTRWTL 149
+ LG + A+ S+ + ++ +S+ A+++ + LA+ + ++ +
Sbjct 234 IESPAPLLGDSVKAIVQFSLEVSCNQNLESSTRHQAIQIVSWLAKYKYNSLKKHKLVIPI 293
Query 150 LNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAA-ICLGLCAQVVKDAILPLVIN 208
L +M PLL E +D P A+A + L + K LP V+
Sbjct 294 LQVM-------CPLLAESSDQEDDD-----DLAPDRASAEVIDTLAMNLPKHVFLP-VLE 340
Query 209 FVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSA 268
F ++ ST + REASV ALG + EG + M + I++ A+ D + VR +A
Sbjct 341 FASVHCQSTNLKFREASVTALGVISEG-CFDLMKEKLDTVLNIVLGAL-RDPELVVRGAA 398
Query 269 AWTLGKTAQQHAPIVLRH 286
++ +G+ A+ P +L H
Sbjct 399 SFAIGQFAEHLQPEILSH 416
> At2g16960
Length=547
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 52/109 (47%), Gaps = 11/109 (10%)
Query 171 AAEDED----DDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLN---FGSTEWRRRE 223
A ED+D +++ W +A +G+ A V D IL ++ ++ F W+ RE
Sbjct 2 AYEDDDETLLNEEEWNLRACSAKFIGILANVFGDEILLTLMPLIEAKLSKFDDETWKERE 61
Query 224 ASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTL 272
A+V A G + EG ++ P++ + ++ + D S VR WTL
Sbjct 62 AAVFAFGAIAEGCNS-FFYPHLIVA---ILRRLLDDQSPLVRRITCWTL 106
> CE26971
Length=1092
Score = 40.0 bits (92), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 72/316 (22%), Positives = 128/316 (40%), Gaps = 60/316 (18%)
Query 32 PDPQLKVAALRALYHALIFAKENFAKTEERDYI------ISSVLEAGNAANPSAVQVAAL 85
PD Q+K A+RA+ + FA +N EE+D + + +VL+ N + L
Sbjct 179 PDLQIKATAVRAV---IAFAVDN---DEEKDVVRLMTSLVPNVLQVCNETSDEDDSDGPL 232
Query 86 ECIVQISQEYYSFLGAYMPAVGSVTWN--ALKSSDSAVVIAALEVWNALAEEELAIIEGE 143
+++ L +M V VT K + V A+EV + E + +G
Sbjct 233 GEFAELASSLPKCLNTHMSQVLQVTLAIAGNKEKNEMVRQNAIEVICSYME---SAPKG- 288
Query 144 STRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDD--DSW------------TPAMAAAI 189
+K+ P L +LE LL+ + DDD + W P +A +
Sbjct 289 ---------LKKYAPGALGPILETLLSCMTEMDDDVLNEWLNEIEEEEDYEDIPIIAESA 339
Query 190 CLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSF 249
+ + +LP+ + V+ S +W+ + A++ A V EG +M P++ +
Sbjct 340 IDRVACCINGKVMLPVFLPLVEKLLTSEDWKMKHAALRAFSAVGEG-CQRSMEPHIEQIM 398
Query 250 PILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHLFPPADVSSTTSPNNNGGALLLA 309
+ V +DA V+ +A +G+ + AP + + A++ A
Sbjct 399 AHITNYV-NDAHPRVQYAACNAIGQMSSDFAPTLQKKCH---------------AAVIPA 442
Query 310 IVERL--TDQPRVAVH 323
++E L TD PRV H
Sbjct 443 LLESLDRTDVPRVCAH 458
> HsM11545781
Length=774
Score = 40.0 bits (92), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 48/210 (22%), Positives = 96/210 (45%), Gaps = 13/210 (6%)
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
AG+ PS +++ AL+ + +++ Y+S AY+ +G V + +D ++ + ++
Sbjct 271 AGSTYEPSPMRLEALQVLTLLARGYFSMTQAYLMELGEVICKCMGEADPSIQLHGAKLLE 330
Query 131 ALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAAIC 190
L + + +ST + A FL+ + +LN +S P + A+ C
Sbjct 331 ELGTGLIQQYKPDSTAAP----DQRAPVFLVVMFWTMMLNGPLPRALQNSEHPTLQASAC 386
Query 191 LGLCAQVVKDAI--LP----LVINFVQLNFGSTEWRR-REASVLALGCVMEGPSAEAMAP 243
L + ++ +A LP ++ V L ++ R + A+ ALG + P
Sbjct 387 DAL-SSILPEAFSNLPNDRQMLCITVLLGLNDSKNRLVKAATSRALGVYVLFPCLRQDVI 445
Query 244 YVARSFPILVEAVASDASVAVRDSAAWTLG 273
+VA + ++ ++ D S+ VR AAW+LG
Sbjct 446 FVADAANAILMSL-EDKSLNVRAKAAWSLG 474
> Hs21361131
Length=1097
Score = 38.5 bits (88), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 63/284 (22%), Positives = 117/284 (41%), Gaps = 35/284 (12%)
Query 15 QQRCNVVLTAVAHGLKDPD-PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN 73
Q +V+ + ++D + P ++ + RA ++ + N A + ++ L+A
Sbjct 167 QHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNVALFKHFADLLPGFLQA-- 224
Query 74 AANPSAVQV--AALECIVQISQEYYSFLGAYMPAV--------GSVTWNALKSSDSAVVI 123
N S Q + L+ +V+I+ +L ++ A G + N ++
Sbjct 225 -VNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTSLNNMQRQ------ 277
Query 124 AALEVWNALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD 178
ALEV L+E A++ + NI+ + P +L +++ E NA E EDDD
Sbjct 278 LALEVIVTLSETAAAMLRKHT------NIVAQTIPQMLAMMVDLEEDEDWANADELEDDD 331
Query 179 -DSWTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPS 237
DS A +A+ C K +LP++ + + +W+ R A ++AL + EG
Sbjct 332 FDSNAVAGESALDRMACGLGGK-LVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCH 390
Query 238 AEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
+ + V D VR +A +G+ A AP
Sbjct 391 QQMEG--ILNEIVNFVLLFLQDPHPRVRYAACNAVGQMATDFAP 432
> CE18111
Length=883
Score = 38.1 bits (87), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Query 181 WTPAMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEA 240
W +A L + A + +L + ++ + W +E+ +LALG + EG +
Sbjct 363 WNIRRCSAASLDVLASIFGKDLLDKLFPLLKDTLMNDNWLVKESGILALGAIAEG-CMDG 421
Query 241 MAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHA 280
+ P++ P ++ A+ D VR WTL + + A
Sbjct 422 VVPHLGELIPFML-AMMFDKKPLVRSITCWTLSRYSSHIA 460
> Hs21361897
Length=1013
Score = 37.4 bits (85), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 48/211 (22%), Positives = 95/211 (45%), Gaps = 15/211 (7%)
Query 71 AGNAANPSAVQVAALECIVQISQEYYSFLGAYMPAVGSVTWNALKSSDSAVVIAALEVWN 130
AG+ PS +++ AL+ + +++ Y+S AY+ +G V + +D ++ ++
Sbjct 510 AGSTYEPSPMRLEALQVLTLLARGYFSMTQAYLMELGEVICKCMGEADPSIQPHGAKLLE 569
Query 131 ALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-EQLLNAAEDEDDDDSWTPAMAAAI 189
L + + +ST + P LL ++ +LN +S P + A+
Sbjct 570 ELGTGLIQQYKPDST-----TAPDQRAPVLLVVMFWTMMLNGPLPRALQNSEHPTLQASA 624
Query 190 CLGLCAQVVKDAI--LP----LVINFVQLNFGSTEWRR-REASVLALGCVMEGPSAEAMA 242
C L + ++ +A LP ++ V L ++ R + A+ ALG + P
Sbjct 625 CDAL-SSILPEAFSNLPNDRQMLCITVLLGLNDSKNRLVKAATSRALGVYVLFPCLRQDV 683
Query 243 PYVARSFPILVEAVASDASVAVRDSAAWTLG 273
+VA + ++ ++ D S+ VR AAW+LG
Sbjct 684 IFVADAANAILMSL-EDKSLNVRAKAAWSLG 713
> HsM4504909
Length=1097
Score = 36.6 bits (83), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 61/278 (21%), Positives = 116/278 (41%), Gaps = 23/278 (8%)
Query 15 QQRCNVVLTAVAHGLKDPD-PQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGN 73
Q +V+ + ++D + P ++ + RA ++ + N A + ++ L+A
Sbjct 167 QHYLDVIKRMLVQCMQDQEHPSIRTLSARATAAFILANEHNVALFKHFADLLPGFLQA-- 224
Query 74 AANPSAVQV--AALECIVQISQEYYSFLGAYMPAVGSVTWNAL--KSSDSAVVIAALEVW 129
N S Q + L+ +V+I+ +L ++ A ++ S ++ ALEV
Sbjct 225 -VNDSCYQNDDSVLKSLVEIADTVPKYLRPHLEATLQLSLKLCGDTSXNNMQRQLALEVI 283
Query 130 NALAEEELAIIEGESTRWTLLNIMKEATPFLLPLLL-----EQLLNAAEDEDDD-DSWTP 183
+E A++ + NI+ + P +L +++ E NA E EDDD DS
Sbjct 284 VTXSETAAAMLRKHT------NIVAQTIPQMLAMMVDLEEDEDWANADELEDDDFDSNAV 337
Query 184 AMAAAICLGLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAP 243
A +A+ C K +LP++ + + +W+ R A ++AL + EG +
Sbjct 338 AGESALDRMACGLGGK-LVLPMIKEHIMQMLQNPDWKYRHAGLMALSAIGEGCHQQMEG- 395
Query 244 YVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
+ V D VR +A +G+ A AP
Sbjct 396 -ILNEIVNFVLLFLQDPHPRVRYAACNAVGQMATDFAP 432
> CE27001
Length=938
Score = 33.9 bits (76), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 61/149 (40%), Gaps = 18/149 (12%)
Query 86 ECIVQISQEYYSFLGAYMPAVGSVTWNALKSSD-----SAVVIAALEVWNALAEEELAII 140
E SQ Y + ++P + WN LKS+ +V AALE + +++ +
Sbjct 267 EIFTLYSQRYEEEISEFVPDIILAVWNLLKSTGPDTRYDTMVCAALEFLSMVSQRQY--Y 324
Query 141 EGESTRWTLLNIMKEATPFLLPLLLEQLLNAAEDEDDD---------DSWTPAMAAA-IC 190
EG T +L + E LL +Q + EDE D D T A +
Sbjct 325 EGHFTGEGVLKTLAENVCVQNLLLRQQDMELFEDEPLDYMKRDIEGTDVGTRRRGAIDLA 384
Query 191 LGLCAQVVKDAILPLVINFVQLNFGSTEW 219
GLC + + +LP + VQ GS +W
Sbjct 385 RGLCRR-FEAQMLPCLGEIVQNLLGSGDW 412
> At1g72440
Length=1056
Score = 33.5 bits (75), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query 15 QQRCNVVLTAVAHGLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGNA 74
+QR +TA+ KD P+LK AL+ +Y L E +ER ++S V + G+
Sbjct 315 KQRYERFVTALDESSKDMLPELKDKALKTIYFMLTSKSE-----QERKLLVSLVNKLGDP 369
Query 75 ANPSA 79
N SA
Sbjct 370 QNKSA 374
> YIL128w
Length=1032
Score = 33.5 bits (75), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 228 ALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIVLRHL 287
AL V++ ++++ P++ FP+L++A+ VR SA TL T +H ++ H+
Sbjct 891 ALSLVLKHTPSQSVGPFINDLFPLLLQALDM-PDPEVRVSALETLKDTTDKHHTLITEHV 949
> At1g50030
Length=2467
Score = 33.5 bits (75), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 240 AMAPYVARSFPILVEAVASDASVAVRDSAAWTLGKTAQQHAPIV 283
AM Y+ P++VEA+ A+VA R+ A TLG+ Q +V
Sbjct 715 AMRQYIPELMPLIVEALMDGAAVAKREVAVSTLGQVVQSTGYVV 758
> At1g47380
Length=439
Score = 33.5 bits (75), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 5/103 (4%)
Query 59 EERDYIISSVLEAGNAANPSAVQVAALEC---IVQISQEYYSF-LGAYMPAVGSVTWNAL 114
EERD + +S E G ++ L C + +S+ +G Y+ V V L
Sbjct 174 EERDRVTASGGEVGRLNTGGGTEIGPLRCWPGGLCLSRSIGDLDVGEYIVPVPYVKQVKL 233
Query 115 KSSDSAVVIAALEVWNAL-AEEELAIIEGESTRWTLLNIMKEA 156
S+ ++I++ VW+A+ AEE L G + +I+KEA
Sbjct 234 SSAGGRLIISSDGVWDAISAEEALDCCRGLPPESSAEHIVKEA 276
> SPCC1840.03
Length=1095
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 62/270 (22%), Positives = 114/270 (42%), Gaps = 24/270 (8%)
Query 28 GLKDPDPQLKVAALRALYHALIFAKENFAKTEERDYIISSVLEAGNAANPSAVQVAA--- 84
GL DP +++++A RA Y A+I +K RD +I + N P +
Sbjct 184 GLADPSIRVRISAARA-YSAVILE----SKQSTRDQVIPLLPSLMNILPPLQQDRDSDNL 238
Query 85 LECIVQISQEYYSFLGAYMPAVGSVTWNAL-----KSSDSAVVIAALEVWNALAEEELAI 139
+C++ I++ F + P SV L K D++ AALE+ +E A+
Sbjct 239 ADCLMAITEIAEVFPKLFKPIFESVIAFGLGIIKDKELDNSARQAALELLVCFSEGAPAM 298
Query 140 IEGESTRWT--------LLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAAICL 191
+S+ +T LL P L++ LN + + D+ +A
Sbjct 299 CR-KSSDYTDQLVLQCLLLMTDVAGDPEDEAEELQEWLNTDDLDQDESDANHVVAEQAMD 357
Query 192 GLCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPI 251
L ++ ILP ++ S +W R A+++A+ + EG + + M ++R +
Sbjct 358 RLSRKLGGKTILPPSFTWLPRLIPSQKWSERHAALMAISSIAEG-AEKLMKKELSRVLDM 416
Query 252 LVEAVASDASVAVRDSAAWTLGKTAQQHAP 281
++ +A D VR +A +G+ + AP
Sbjct 417 VLPLLA-DPHPRVRWAACNAVGQMSTDFAP 445
> At1g64790
Length=2428
Score = 31.2 bits (69), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 29/131 (22%), Positives = 56/131 (42%), Gaps = 12/131 (9%)
Query 153 MKEATPFLLPLLLEQLLNAAEDEDDDDSWTPAMAAAICLGLCAQVVKDAILPLVINFVQL 212
+KE P L+ L+ L + + + A LG + + + +LPL+I +
Sbjct 1679 LKEIMPILMSTLISSLASPSSERRQ--------VAGRSLGELVRKLGERVLPLIIPILSK 1730
Query 213 NFGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAW-- 270
+ +R+ + L VM + ++ + P + A+ D+++ VR+SA
Sbjct 1731 GLKDPDVDKRQGVCIGLNEVMASAGRSQLLSFMDQLIPTIRTALC-DSALEVRESAGLAF 1789
Query 271 -TLGKTAQQHA 280
TL K+A A
Sbjct 1790 STLYKSAGLQA 1800
> 7294673
Length=1876
Score = 30.8 bits (68), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 29/69 (42%), Gaps = 1/69 (1%)
Query 288 FPPADVSSTTSPNNNGGALLLAIVERLTDQPRVAVHICWML-HELADDINTPEGRAAYQG 346
F P V S G A+ ++ + + PR A H+ W+L H L DD + G A
Sbjct 1143 FLPQLVQSLKHDTYEGSAMARFLLSKCLESPRFAHHMYWLLVHSLPDDPHNSIGAAMVDQ 1202
Query 347 EAATQQQDQ 355
E Q Q
Sbjct 1203 EYDESQVTQ 1211
> Hs6912678
Length=509
Score = 30.8 bits (68), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 1/68 (1%)
Query 214 FGSTEWRRREASVLALGCVMEGPSAEAMAPYVARSFPILVEAVASDASVAVRDSAAWTLG 273
GS + R ++ LG V AMA +++ P L ++ + ++ +AAW LG
Sbjct 302 IGSCKGNTRLPGIMMLGYVAAHSENLAMAVIISKGVPQLSVCLSEEPEDHIKAAAAWALG 361
Query 274 KTAQQHAP 281
+ +H P
Sbjct 362 QIG-RHTP 368
> At5g19820
Length=1116
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 53/225 (23%), Positives = 93/225 (41%), Gaps = 39/225 (17%)
Query 36 LKVAALRALYHALIFAKENFAKTEERDY-------IISSVLEAGNAANPSAVQVAALECI 88
+K+AAL +A+I + A + ERD +I ++ E+ N N + Q ALE +
Sbjct 202 VKIAAL----NAVISFVQCLANSTERDRFQDVLPAMIRTLTESLNNGNEATAQ-EALELL 256
Query 89 VQISQEYYSFLGAYM-PAVGSVTWNALKSS-DSAVVIAALEVWNALAEEELAIIEGESTR 146
++++ FL + VGS+ A S + + A+E LAE R
Sbjct 257 IELAGTEPRFLRRQLVDIVGSMLQIAEADSLEESTRHLAIEFLVTLAE----------AR 306
Query 147 WTLLNIMKEATPFLLPLLLEQLLNAAEDEDDDDSWTPA--------------MAAAICLG 192
++++ P + L L+ ED +DD +W A M
Sbjct 307 ERAPGMVRK-LPQFIDRLFAVLMKMLEDIEDDPAWYSAETEDEDAGETSNYSMGQECLDR 365
Query 193 LCAQVVKDAILPLVINFVQLNFGSTEWRRREASVLALGCVMEGPS 237
L + + I+P+ ++EW++ AS++AL + EG S
Sbjct 366 LAISLGGNTIVPVAYQQFSAYLAASEWQKHHASLIALAQIAEGCS 410
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8980297216
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40