bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0974_orf1
Length=247
Score E
Sequences producing significant alignments: (Bits) Value
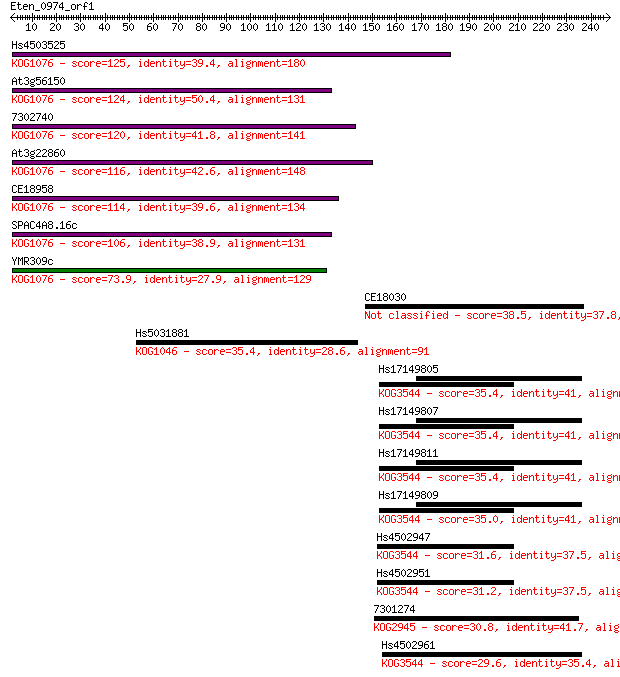
Hs4503525 125 1e-28
At3g56150 124 2e-28
7302740 120 3e-27
At3g22860 116 4e-26
CE18958 114 2e-25
SPAC4A8.16c 106 4e-23
YMR309c 73.9 3e-13
CE18030 38.5 0.014
Hs5031881 35.4 0.10
Hs17149805 35.4 0.12
Hs17149807 35.4 0.12
Hs17149811 35.4 0.12
Hs17149809 35.0 0.14
Hs4502947 31.6 1.7
Hs4502951 31.2 2.3
7301274 30.8 2.7
Hs4502961 29.6 5.3
> Hs4503525
Length=913
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 108/184 (58%), Gaps = 21/184 (11%)
Query 2 VMAATKALQRGDWSDCARHIWSLS----IWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA++ GDW C I + +WD + ++ ML++KI+ E++RTY+FTY
Sbjct 737 VVAASKAMKMGDWKTCHSFIINEKMNGKVWDLFPEADKVRTMLVRKIQEESLRTYLFTYS 796
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
S+YDS ++ L+ MF+L L VHSI+SKM+INEE+ AS D+ +Q +++ R + T Q LA
Sbjct 797 SVYDSISMETLSDMFELDLPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTAQQNLA 856
Query 118 LTLAENVANAVEQNELTLNMKNPKFALTHERRFMRDGDRAGWGTRDREDGGGHRSRPGFR 177
L LAE + + VE NE + K G G+ RD++D G+R G+
Sbjct 857 LQLAEKLGSLVENNERVFDHKQ--------------GTYGGY-FRDQKD--GYRKNEGYM 899
Query 178 GRAG 181
R G
Sbjct 900 RRGG 903
> At3g56150
Length=900
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 91/131 (69%), Gaps = 0/131 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
VMAAT+AL +GD+ + SL +W + N++ I M+ +IK EA+RTY+FTY S Y+
Sbjct 668 VMAATRALTKGDFQKAFEVLNSLEVWRLLKNRDSILDMVKDRIKEEALRTYLFTYSSSYE 727
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTLA 121
S ++ QLA MFD+S VHSIVSKMMINEE+HASWD+ ++ I+ V+H+RLQ LA L
Sbjct 728 SLSLDQLAKMFDVSEPQVHSIVSKMMINEELHASWDQPTRCIVFHEVQHSRLQSLAFQLT 787
Query 122 ENVANAVEQNE 132
E ++ E NE
Sbjct 788 EKLSILAESNE 798
> 7302740
Length=910
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 59/145 (40%), Positives = 93/145 (64%), Gaps = 4/145 (2%)
Query 2 VMAATKALQRGDWSDCARHI----WSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA KA++ G+W CA I + +WD + + +++ML + IK E++RTY+FTY
Sbjct 703 VVAAAKAMRCGNWQACANFIVNKKMNTKVWDLFYESDRVREMLTKFIKEESLRTYLFTYS 762
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
++Y S +I LA M++L + VHSI+SKM+INEE+ AS D+ S+ + + R + +RLQ LA
Sbjct 763 NVYTSISIPSLAQMYELPVPKVHSIISKMIINEELMASLDDPSETVGMHRSEPSRLQALA 822
Query 118 LTLAENVANAVEQNELTLNMKNPKF 142
+ + V N V+ NE +MK F
Sbjct 823 MQFVDKVTNLVDVNEKVFDMKQGNF 847
> At3g22860
Length=800
Score = 116 bits (291), Expect = 4e-26, Method: Composition-based stats.
Identities = 63/151 (41%), Positives = 92/151 (60%), Gaps = 3/151 (1%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLS-LY 60
V+AAT+AL +G++ + + SL IW N++ I M+ I A+RTY+FTY S Y
Sbjct 621 VIAATRALIKGNFQEAFSVLNSLDIWRLFKNRDSILDMVKASISEVALRTYLFTYSSSCY 680
Query 61 DSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTL 120
S ++ +LA MFD+S + V+SIVSKMMIN+E++A+WD+ +Q I+ V+H R Q LA +
Sbjct 681 KSLSLAELAKMFDISESHVYSIVSKMMINKELNATWDQPTQCIVFHEVQHNRAQSLAFQI 740
Query 121 AENVANAVEQNELTLNMKNPKFAL--THERR 149
E +A E NE + K L T RR
Sbjct 741 TEKLATLAESNESAMESKTGGVGLDMTSRRR 771
> CE18958
Length=898
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 53/138 (38%), Positives = 91/138 (65%), Gaps = 4/138 (2%)
Query 2 VMAATKALQRGDWSDCARHI----WSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYL 57
V+AA+KA+ GDW C +I + +W+ N E ++ M++++I+ E++RTY+ TY
Sbjct 694 VVAASKAMLNGDWKKCQDYIVNDKMNQKVWNLFHNAETVKGMVVRRIQEESLRTYLLTYS 753
Query 58 SLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLA 117
++Y + ++ +LA +F+LS VHSI+SKM+I EE+ A+ DE + +++ RV+ +RLQ LA
Sbjct 754 TVYATVSLKKLADLFELSKKDVHSIISKMIIQEELSATLDEPTDCLIMHRVEPSRLQMLA 813
Query 118 LTLAENVANAVEQNELTL 135
L L++ + E NE L
Sbjct 814 LNLSDKLQTLAENNEQIL 831
> SPAC4A8.16c
Length=639
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/131 (38%), Positives = 80/131 (61%), Gaps = 0/131 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
+M A+KAL G+W C I ++ IW M + + I+QML +KI+ E +RTY+ Y + YD
Sbjct 469 IMQASKALADGEWRRCEEFIHAIKIWSLMPDADKIKQMLSEKIREEGLRTYLLAYAAFYD 528
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTRLQQLALTLA 121
S ++ LA+ FDL + V IVS+++ EIHA+ D+ I+ RV+ +L+ L ++L+
Sbjct 529 SVSLEFLATTFDLPVQRVTVIVSRLLSKREIHAALDQVHGAIIFERVEINKLESLTVSLS 588
Query 122 ENVANAVEQNE 132
E A E NE
Sbjct 589 EKTAQLNEANE 599
> YMR309c
Length=812
Score = 73.9 bits (180), Expect = 3e-13, Method: Composition-based stats.
Identities = 36/130 (27%), Positives = 74/130 (56%), Gaps = 1/130 (0%)
Query 2 VMAATKALQRGDWSDCARHIWSLSIWDKMFNKEHIQQMLLQKIKAEAMRTYIFTYLSLYD 61
V+ A K++Q+G+W D +++ + W + N E + L ++++ E+++TY F++ Y
Sbjct 675 VLFAAKSMQKGNWRDSVKYLREIKSWALLPNMETVLNSLTERVQVESLKTYFFSFKRFYS 734
Query 62 SFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASW-DETSQFILISRVKHTRLQQLALTL 120
SF++ +LA +FDL N V ++ ++ EI A DE + F++ + T+L++ + L
Sbjct 735 SFSVAKLAELFDLPENKVVEVLQSVIAELEIPAKLNDEKTIFVVEKGDEITKLEEAMVKL 794
Query 121 AENVANAVEQ 130
+ A E+
Sbjct 795 NKEYKIAKER 804
> CE18030
Length=798
Score = 38.5 bits (88), Expect = 0.014, Method: Composition-based stats.
Identities = 34/96 (35%), Positives = 45/96 (46%), Gaps = 15/96 (15%)
Query 147 ERRFMRDGDRAGWGTRDREDGGGHRSRPGFRGRA-GLNPVGARGRGLGRGTYRGCGFRGN 205
+R R GDR G R + G +R R F G + G N RG G G T RG
Sbjct 108 DRGNFRGGDRGGSPWRGGDRGVANRGRGDFSGGSRGGNKFSPRGGGRGGFTPRG------ 161
Query 206 RGSGDY-----RGDYCGESRGDLGEYRGYSDQRDPF 236
RG D+ RG++ +SRG G G+SD+R +
Sbjct 162 RGGNDFSPRGGRGNF--QSRGG-GSRVGFSDKRKQY 194
> Hs5031881
Length=1025
Score = 35.4 bits (80), Expect = 0.10, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 41/91 (45%), Gaps = 9/91 (9%)
Query 53 IFTYLSLYDSFAIGQLASMFDLSLNAVHSIVSKMMINEEIHASWDETSQFILISRVKHTR 112
IF LS Y+ F + +M SLN+ H I S + +E+I +D S F +
Sbjct 501 IFKELSSYEDFLDARFKTMKKDSLNSSHPISSSVQSSEQIEEMFDSLSYF---------K 551
Query 113 LQQLALTLAENVANAVEQNELTLNMKNPKFA 143
L L L ++ V Q+ + L + N +A
Sbjct 552 GSSLLLMLKTYLSEDVFQHAVVLYLHNHSYA 582
> Hs17149805
Length=3009
Score = 35.4 bits (80), Expect = 0.12, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 168 GGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG-SGDYRGDYCGES--RGDLG 224
GG R PG RGR G P+G +G G G G G RG +GD D G RG G
Sbjct 2087 GGARGAPGERGRTG--PLGRKGEPGEPGPKGGIGNPGPRGETGDDGRDGVGSEGRRGKKG 2144
Query 225 EYRGYSDQRDP 235
E RG+ P
Sbjct 2145 E-RGFPGYPGP 2154
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 32/77 (41%), Gaps = 22/77 (28%)
Query 153 DGDRAGWGTRDREDGGGHRSRPGF------RGRAGLN----PVGARGR-------GL--- 192
D R G G+ R G R PG+ G GLN P G RGR G+
Sbjct 2128 DDGRDGVGSEGRRGKKGERGFPGYPGPKGNPGEPGLNGTTGPKGIRGRRGNSGPPGIVGQ 2187
Query 193 -GRGTYRG-CGFRGNRG 207
GR Y G G RGNRG
Sbjct 2188 KGRPGYPGPAGPRGNRG 2204
> Hs17149807
Length=2976
Score = 35.4 bits (80), Expect = 0.12, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 168 GGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG-SGDYRGDYCGES--RGDLG 224
GG R PG RGR G P+G +G G G G G RG +GD D G RG G
Sbjct 2054 GGARGAPGERGRTG--PLGRKGEPGEPGPKGGIGNPGPRGETGDDGRDGVGSEGRRGKKG 2111
Query 225 EYRGYSDQRDP 235
E RG+ P
Sbjct 2112 E-RGFPGYPGP 2121
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 32/77 (41%), Gaps = 22/77 (28%)
Query 153 DGDRAGWGTRDREDGGGHRSRPGF------RGRAGLN----PVGARGR-------GL--- 192
D R G G+ R G R PG+ G GLN P G RGR G+
Sbjct 2095 DDGRDGVGSEGRRGKKGERGFPGYPGPKGNPGEPGLNGTTGPKGIRGRRGNSGPPGIVGQ 2154
Query 193 -GRGTYRG-CGFRGNRG 207
GR Y G G RGNRG
Sbjct 2155 KGRPGYPGPAGPRGNRG 2171
> Hs17149811
Length=2970
Score = 35.4 bits (80), Expect = 0.12, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 168 GGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG-SGDYRGDYCGES--RGDLG 224
GG R PG RGR G P+G +G G G G G RG +GD D G RG G
Sbjct 2048 GGARGAPGERGRTG--PLGRKGEPGEPGPKGGIGNPGPRGETGDDGRDGVGSEGRRGKKG 2105
Query 225 EYRGYSDQRDP 235
E RG+ P
Sbjct 2106 E-RGFPGYPGP 2115
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 32/77 (41%), Gaps = 22/77 (28%)
Query 153 DGDRAGWGTRDREDGGGHRSRPGF------RGRAGLN----PVGARGR-------GL--- 192
D R G G+ R G R PG+ G GLN P G RGR G+
Sbjct 2089 DDGRDGVGSEGRRGKKGERGFPGYPGPKGNPGEPGLNGTTGPKGIRGRRGNSGPPGIVGQ 2148
Query 193 -GRGTYRG-CGFRGNRG 207
GR Y G G RGNRG
Sbjct 2149 KGRPGYPGPAGPRGNRG 2165
> Hs17149809
Length=2975
Score = 35.0 bits (79), Expect = 0.14, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 168 GGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG-SGDYRGDYCGES--RGDLG 224
GG R PG RGR G P+G +G G G G G RG +GD D G RG G
Sbjct 2053 GGARGAPGERGRTG--PLGRKGEPGEPGPKGGIGNPGPRGETGDDGRDGVGSEGRRGKKG 2110
Query 225 EYRGYSDQRDP 235
E RG+ P
Sbjct 2111 E-RGFPGYPGP 2120
Score = 32.0 bits (71), Expect = 1.2, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 32/77 (41%), Gaps = 22/77 (28%)
Query 153 DGDRAGWGTRDREDGGGHRSRPGF------RGRAGLN----PVGARGR-------GL--- 192
D R G G+ R G R PG+ G GLN P G RGR G+
Sbjct 2094 DDGRDGVGSEGRRGKKGERGFPGYPGPKGNPGEPGLNGTTGPKGIRGRRGNSGPPGIVGQ 2153
Query 193 -GRGTYRG-CGFRGNRG 207
GR Y G G RGNRG
Sbjct 2154 KGRPGYPGPAGPRGNRG 2170
> Hs4502947
Length=1366
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 2/56 (3%)
Query 152 RDGDRAGWGTRDREDGGGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFRGNRG 207
RDG+ G R+ GH+ G+ G G PVGA G G G GNRG
Sbjct 926 RDGNPGNDGPPGRDGQPGHKGERGYPGNIG--PVGAAGAPGPHGPVGPAGKHGNRG 979
> Hs4502951
Length=1466
Score = 31.2 bits (69), Expect = 2.3, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 24/57 (42%), Gaps = 1/57 (1%)
Query 152 RDGDRAGWGTRDREDGGGHRSRPGFRGRAGLNPV-GARGRGLGRGTYRGCGFRGNRG 207
+DG G G R PGFRG AG N + G +G RG G RG G
Sbjct 466 KDGSPGEPGANGLPGAAGERGAPGFRGPAGPNGIPGEKGPAGERGAPGPAGPRGAAG 522
> 7301274
Length=423
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 44/93 (47%), Gaps = 16/93 (17%)
Query 151 MRDGDRAGWGTRD---REDG----GGHRSRPGFRGRAGLNPVGARGRGLGRGTYRGCGFR 203
R G R G G R+ R DG GG+R+ G RG G G RG G RG G+R
Sbjct 311 FRKGPRPGAGPREGGFRNDGPRGEGGYRND-GPRGEGGYRNDGPRGEGPRNEGPRGEGYR 369
Query 204 --GNRGSGDYRGDYCGESRGDLGEYRGYSDQRD 234
G RG G +RG R + G GY +++D
Sbjct 370 NDGPRGEG-FRG-----PRFNNGPSNGYENRQD 396
> Hs4502961
Length=2944
Score = 29.6 bits (65), Expect = 5.3, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 11/87 (12%)
Query 154 GDRAGWGTRDREDGGGHRSRPGFRGRAGLN-PVGARGRGLGRGTYRGCGFRGNRGSGDYR 212
G+R G R EDG RPG G GL P G+RG +G G +G++G
Sbjct 2472 GERGEPGIRG-EDG-----RPGQEGPRGLTGPPGSRGERGEKGDVGSAGLKGDKGDSAVI 2525
Query 213 GDYCGE--SRGDLGEY--RGYSDQRDP 235
G ++GD+GE RG + P
Sbjct 2526 LGPPGPRGAKGDMGERGPRGLDGDKGP 2552
Lambda K H
0.321 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5009866206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40