bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0922_orf4
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
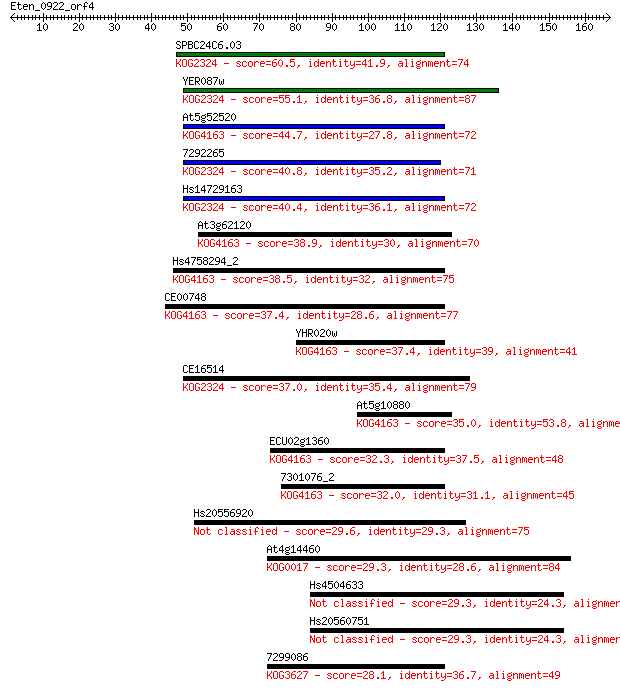
SPBC24C6.03 60.5 1e-09
YER087w 55.1 7e-08
At5g52520 44.7 8e-05
7292265 40.8 0.001
Hs14729163 40.4 0.002
At3g62120 38.9 0.005
Hs4758294_2 38.5 0.007
CE00748 37.4 0.015
YHR020w 37.4 0.015
CE16514 37.0 0.016
At5g10880 35.0 0.073
ECU02g1360 32.3 0.42
7301076_2 32.0 0.62
Hs20556920 29.6 3.1
At4g14460 29.3 3.6
Hs4504633 29.3 3.7
Hs20560751 29.3 3.8
7299086 28.1 7.4
> SPBC24C6.03
Length=425
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 46/74 (62%), Gaps = 1/74 (1%)
Query 47 TCLEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLL 106
T +EVGH F L K Y A ++ + ++V M YG+GVSRLIA +A +D KGL+
Sbjct 263 TAIEVGHAFYLGKIYSSKFNAT-VEVKNKQEVLHMGCYGIGVSRLIAAVAHVTKDAKGLV 321
Query 107 FPPQVAPFCVAIIP 120
+P +AP+ V ++P
Sbjct 322 WPSSIAPWKVLVVP 335
> YER087w
Length=576
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 32/88 (36%), Positives = 46/88 (52%), Gaps = 5/88 (5%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGV-EKVPFMSSYGLGVSRLIAHLALSHQDMKGLLF 107
+EVGH F L Y K +F+D + E M YG+GVSRL+ +A +D G +
Sbjct 379 IEVGHIFLLGNKYSKPLNVKFVDKENKNETFVHMGCYGIGVSRLVGAIAELGRDSNGFRW 438
Query 108 PPQVAPFCVAIIPQPAARWNDGRNAARL 135
P +AP+ V+I P N+ N+ RL
Sbjct 439 PAIMAPYKVSICTGP----NNPENSQRL 462
> At5g52520
Length=543
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFP 108
L+ G L + + +A +F D G + + +S+ + +R + + ++H D GL+ P
Sbjct 283 LQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVS-TRFVGGIIMTHGDDTGLMLP 341
Query 109 PQVAPFCVAIIP 120
P++AP V I+P
Sbjct 342 PKIAPIQVVIVP 353
> 7292265
Length=458
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 8/75 (10%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAH----LALSHQDMKG 104
+EV H F L Y K A F+++ G + M YG+G++R+IA L+ H+
Sbjct 264 VEVAHTFLLGDKYSKPLGATFLNTTGKPQSLVMGCYGIGITRVIAAALEVLSSDHE---- 319
Query 105 LLFPPQVAPFCVAII 119
L +P +AP+ V +I
Sbjct 320 LRWPKLLAPYDVCLI 334
> Hs14729163
Length=402
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHL--ALSHQDMKGLL 106
+EVGH F L Y A+F + G + M YGLGV+R++A LS +D +
Sbjct 232 IEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVLSTEDC--VR 289
Query 107 FPPQVAPFCVAIIP 120
+P +AP+ +IP
Sbjct 290 WPSLLAPYQACLIP 303
> At3g62120
Length=530
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query 53 HCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVA 112
HC L + + K F + + ++ + +S+ +R I + ++H D KGL+ PP+VA
Sbjct 275 HC--LGQNFAKMFEINFENEKAETEMVWQNSWAYS-TRTIGVMIMTHGDDKGLVLPPKVA 331
Query 113 PFCVAIIPQP 122
V +IP P
Sbjct 332 SVQVVVIPVP 341
> Hs4758294_2
Length=499
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 3/77 (3%)
Query 46 GTCLEVGHCFQLPKAYCKAARARFMDSQ--GVEKVPFMSSYGLGVSRLIAHLALSHQDMK 103
G ++ G L + + K F D + G ++ + +S+GL +R I + + H D
Sbjct 220 GRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGL-TTRTIGVMTMVHGDNM 278
Query 104 GLLFPPQVAPFCVAIIP 120
GL+ PP+VA V IIP
Sbjct 279 GLVLPPRVACVQVVIIP 295
> CE00748
Length=581
Score = 37.4 bits (85), Expect = 0.015, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 38/79 (48%), Gaps = 3/79 (3%)
Query 44 CGGTCLEVGHCFQLPKAYCKAARARFMD--SQGVEKVPFMSSYGLGVSRLIAHLALSHQD 101
C G ++ L + + K + D +G + +S+GL +R I + + H D
Sbjct 306 CNGRGIQGATSHHLGQNFSKMFDISYEDPAKEGERAFAWQNSWGLS-TRTIGAMVMIHGD 364
Query 102 MKGLLFPPQVAPFCVAIIP 120
KGL+ PP+VA V ++P
Sbjct 365 DKGLVLPPRVAAVQVIVVP 383
> YHR020w
Length=688
Score = 37.4 bits (85), Expect = 0.015, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 80 FMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+ +S+GL +R+I + + H D KGL+ PP+V+ F +IP
Sbjct 441 YQNSWGLS-TRVIGVMVMIHSDNKGLVIPPRVSQFQSVVIP 480
> CE16514
Length=454
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 49 LEVGHCFQLPKAYCKAARARFMDSQGVEKVPFMSSYGLGVSRLI-AHLALSHQDMKGLLF 107
+E+ H F L Y +A A+F QG K M +G+GV+RL+ A + L K L
Sbjct 262 VEIAHTFHLGTKYSEALGAKF---QG--KPLDMCCFGIGVTRLLPAAIDLLSVSDKALRL 316
Query 108 PPQVAPFCVAIIPQPAARWN 127
P +APF II + + N
Sbjct 317 PRAIAPFDAVIIVKKSLMSN 336
> At5g10880
Length=309
Score = 35.0 bits (79), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 97 LSHQDMKGLLFPPQVAPFCVAIIPQP 122
++H D KGL+FPP+VAP V +I P
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVIHVP 114
> ECU02g1360
Length=520
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 73 QGVEKVPFMSSYGLGVS-RLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+G E F+ G++ R I A+ H D GL+ PP+VA V I+P
Sbjct 271 EGSESSSFVYQNSWGITTRSIGIAAMIHSDNLGLVLPPRVAMTQVVIVP 319
> 7301076_2
Length=586
Score = 32.0 bits (71), Expect = 0.62, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 76 EKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
+K + +S+G+ +R I + + H D +GL+ PP VA ++P
Sbjct 337 KKYVYQNSWGI-TTRTIGVMIMVHADNQGLVLPPHVACIQAIVVP 380
> Hs20556920
Length=525
Score = 29.6 bits (65), Expect = 3.1, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 11/83 (13%)
Query 52 GHCFQLPKAYCKAARARFMDSQGVEKVPF-MSSYGLGVSRLIAHLALSHQD-------MK 103
G+C KA C + S P+ S+Y + ++ + +S+Q ++
Sbjct 61 GNCSSCHKASCGIEECDCLTSNSASSSPYDFSTYFIQKAKSSSRGVVSNQQAAQSTEGVE 120
Query 104 GLLFPPQVAPFCVAIIPQPAARW 126
GLLFPP FCV + Q W
Sbjct 121 GLLFPP---AFCVGDVTQQRDLW 140
> At4g14460
Length=1489
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 3/87 (3%)
Query 72 SQGVEKVPFMSSYGLGVSRLIAHLA--LSHQDMKGLLFPPQVAPFCVAI-IPQPAARWND 128
S+G + P M+ YG I H LS QD ++ F + +P+PAA ++
Sbjct 367 SEGYSQAPMMNPYGSYPMPHITHGGNNLSLQDFTPQQIEQMISQFQAQVQVPEPAASSSN 426
Query 129 GRNAARLGRALFRRLHRTLGSLVYFTS 155
A + F L T G+++ F S
Sbjct 427 PSPLATVSEHGFMALTSTSGTIIPFPS 453
> Hs4504633
Length=578
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query 84 YGLGVSRLIAHLALSHQDMK----GLLFPPQVAPFCVAIIPQPAARWNDGRNAARLGRAL 139
Y + + ++ + +H+ +K LL +V FCV + P A+R N G + +L
Sbjct 167 YEIAIRKVPGNFTFTHKKVKHENFSLLTSGEVGEFCVQVKPSVASRSNKGMWSKEECISL 226
Query 140 FRRLHRTLGSLVYF 153
R+ +++F
Sbjct 227 TRQYFTVTNVIIFF 240
> Hs20560751
Length=578
Score = 29.3 bits (64), Expect = 3.8, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query 84 YGLGVSRLIAHLALSHQDMK----GLLFPPQVAPFCVAIIPQPAARWNDGRNAARLGRAL 139
Y + + ++ + +H+ +K LL +V FCV + P A+R N G + +L
Sbjct 167 YEIAIRKVPGNFTFTHKKVKHENFSLLTSGEVGEFCVQVKPSVASRSNKGMWSKEECISL 226
Query 140 FRRLHRTLGSLVYF 153
R+ +++F
Sbjct 227 TRQYFTVTNVIIFF 240
> 7299086
Length=377
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 22/49 (44%), Gaps = 4/49 (8%)
Query 72 SQGVEKVPFMSSYGLGVSRLIAHLALSHQDMKGLLFPPQVAPFCVAIIP 120
SQ +VP S G H L+ Q + G L PPQVAP +P
Sbjct 37 SQDTNRVPQPSDVG----STANHTILARQLIVGTLLPPQVAPGTWPPVP 81
Lambda K H
0.328 0.138 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2389760076
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40