bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
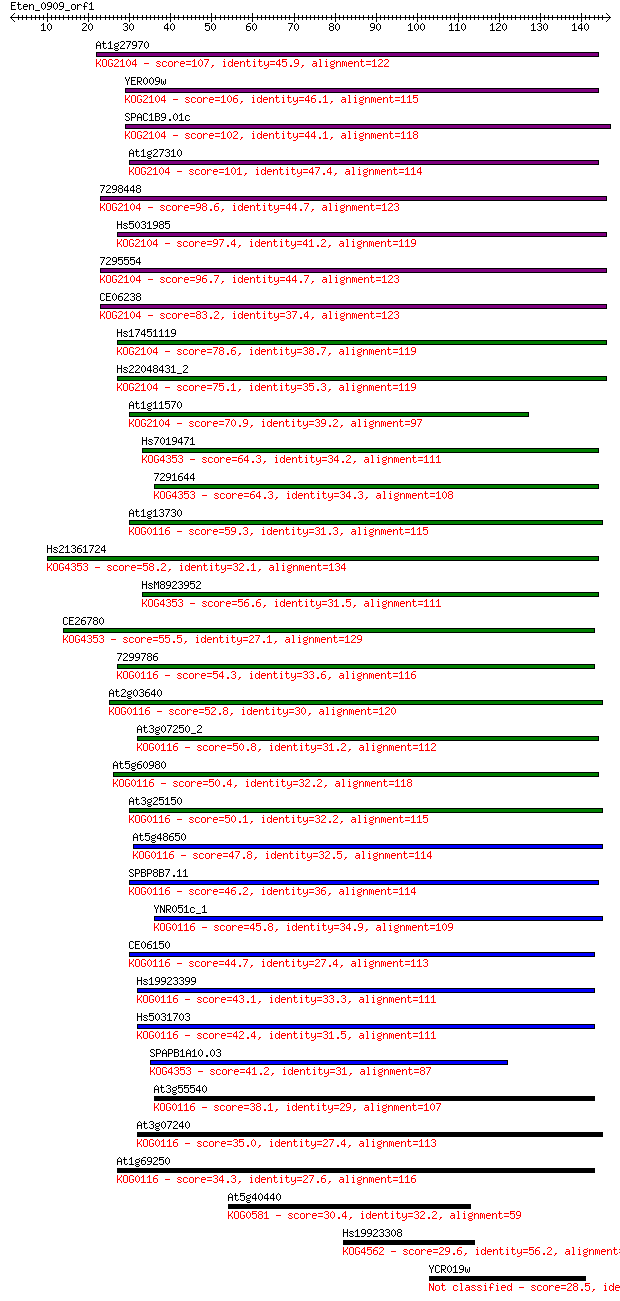
Query= Eten_0909_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
At1g27970 107 9e-24
YER009w 106 2e-23
SPAC1B9.01c 102 2e-22
At1g27310 101 5e-22
7298448 98.6 4e-21
Hs5031985 97.4 9e-21
7295554 96.7 1e-20
CE06238 83.2 2e-16
Hs17451119 78.6 4e-15
Hs22048431_2 75.1 5e-14
At1g11570 70.9 9e-13
Hs7019471 64.3 7e-11
7291644 64.3 7e-11
At1g13730 59.3 3e-09
Hs21361724 58.2 6e-09
HsM8923952 56.6 2e-08
CE26780 55.5 4e-08
7299786 54.3 8e-08
At2g03640 52.8 2e-07
At3g07250_2 50.8 1e-06
At5g60980 50.4 1e-06
At3g25150 50.1 1e-06
At5g48650 47.8 8e-06
SPBP8B7.11 46.2 2e-05
YNR051c_1 45.8 3e-05
CE06150 44.7 6e-05
Hs19923399 43.1 2e-04
Hs5031703 42.4 3e-04
SPAPB1A10.03 41.2 6e-04
At3g55540 38.1 0.006
At3g07240 35.0 0.047
At1g69250 34.3 0.095
At5g40440 30.4 1.3
Hs19923308 29.6 1.9
YCR019w 28.5 4.8
> At1g27970
Length=126
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 56/125 (44%), Positives = 79/125 (63%), Gaps = 5/125 (4%)
Query 22 MLQLNPHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQL 81
M Q++P D++ FV+ YY TF NR GLA LY E S+LT+EG + +G +IV KL L
Sbjct 1 MSQMDP--DAVSKAFVEHYYSTFDTNRVGLAGLYQEASMLTFEGQKIQGVQSIVAKLTSL 58
Query 82 P-AAVSHQLVTCDCQPT-PHGGVLIIVCGDLAIE-DNPPMKFTQTFNLVPNGSGTYAVFN 138
P H + T DCQP+ P G+L+ V G+L + + +KF+Q F+L+P G++ VFN
Sbjct 59 PFQQCKHHISTVDCQPSGPASGMLVFVSGNLQLAGEEHALKFSQMFHLMPTPQGSFYVFN 118
Query 139 DCFRL 143
D FRL
Sbjct 119 DIFRL 123
> YER009w
Length=125
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 53/118 (44%), Positives = 78/118 (66%), Gaps = 4/118 (3%)
Query 29 FDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAVSH 87
F+++ F Q YY F +R+ L LY S+LT+E +Q +GA IV+KL LP V H
Sbjct 5 FNTLAQNFTQFYYNQFDTDRSQLGNLYRNESMLTFETSQLQGAKDIVEKLVSLPFQKVQH 64
Query 88 QLVTCDCQP-TPHGGVLIIVCGDLAI-EDNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
++ T D QP +P+G VL+++ GDL I E+ P +F+Q F+L+P+G+ +Y VFND FRL
Sbjct 65 RITTLDAQPASPNGDVLVMITGDLLIDEEQNPQRFSQVFHLIPDGN-SYYVFNDIFRL 121
> SPAC1B9.01c
Length=123
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 52/121 (42%), Positives = 79/121 (65%), Gaps = 4/121 (3%)
Query 29 FDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAVSH 87
++++ QF Q YY+TF ++R+ L+ LY E S+L++EG Q +G AIV+KL LP V H
Sbjct 4 YNALATQFTQFYYQTFDSDRSQLSSLYREESMLSFEGAQLQGTKAIVEKLVSLPFQRVQH 63
Query 88 QLVTCDCQPT-PHGGVLIIVCGDLAI-EDNPPMKFTQTFNLVPNGSGTYAVFNDCFRLCI 145
++ T D QPT G V+++V G+L + E+ +++Q F+LV N +G Y V ND FRL
Sbjct 64 RISTLDAQPTGTTGSVIVMVTGELLLDEEQMAQRYSQVFHLV-NNNGNYYVLNDLFRLNY 122
Query 146 G 146
G
Sbjct 123 G 123
> At1g27310
Length=122
Score = 101 bits (251), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 54/117 (46%), Positives = 73/117 (62%), Gaps = 4/117 (3%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAVSHQ 88
D++ FV+ YY TF NR GL LY E S+LT+EG + +G+ IV KL LP H
Sbjct 4 DAVAKAFVEHYYSTFDANRPGLVSLYQEGSMLTFEGQKIQGSQNIVAKLTGLPFQQCKHN 63
Query 89 LVTCDCQPT-PHGGVLIIVCGDLAIE-DNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
+ T DCQP+ P GG+L+ V G+L + + +KF+Q F+L+ N G Y VFND FRL
Sbjct 64 ITTVDCQPSGPAGGMLVFVSGNLQLAGEQHALKFSQMFHLISN-QGNYYVFNDIFRL 119
> 7298448
Length=130
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 55/127 (43%), Positives = 76/127 (59%), Gaps = 5/127 (3%)
Query 23 LQLNPHFDSIGPQFVQVYYETFRN--NRAGLAELYTE-NSLLTYEGTQFRGAAAIVQKLQ 79
+ LN ++ IG +FVQ YY F + NR + Y +S +T+EG Q +GA I++K+Q
Sbjct 1 MSLNLQYEDIGKEFVQQYYAIFDDPANRENVINFYNATDSFMTFEGNQIQGAPKILEKVQ 60
Query 80 QLP-AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFN 138
L ++ + T D QPT GGVLIIV G L +D+PP F+Q F L PNG G+ V +
Sbjct 61 SLSFQKIARVITTVDSQPTSDGGVLIIVLGRLKCDDDPPHAFSQIFLLKPNG-GSLFVAH 119
Query 139 DCFRLCI 145
D FRL I
Sbjct 120 DIFRLNI 126
> Hs5031985
Length=127
Score = 97.4 bits (241), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 49/120 (40%), Positives = 71/120 (59%), Gaps = 2/120 (1%)
Query 27 PHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAV 85
P ++ IG F+Q YY+ F N+R L +Y + S LT+EG QF+G AAIV+KL LP +
Sbjct 5 PIWEQIGSSFIQHYYQLFDNDRTQLGAIYIDASCLTWEGQQFQGKAAIVEKLSSLPFQKI 64
Query 86 SHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRLCI 145
H + D QPTP ++ +V G L +++P M F Q F L+ N + + ND FRL +
Sbjct 65 QHSITAQDHQPTPDSCIISMVVGQLKADEDPIMGFHQMF-LLKNINDAWVCTNDMFRLAL 123
> 7295554
Length=130
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 55/127 (43%), Positives = 77/127 (60%), Gaps = 5/127 (3%)
Query 23 LQLNPHFDSIGPQFVQVYYETFRN--NRAGLAELYTE-NSLLTYEGTQFRGAAAIVQKLQ 79
+ LNP ++ IG FVQ YY F + NRA + Y+ +S +T+EG Q +GA I++K+Q
Sbjct 1 MSLNPQYEDIGKGFVQQYYAIFDDPANRANVVNFYSATDSFMTFEGHQIQGAPKILEKVQ 60
Query 80 QLP-AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFN 138
L ++ + T D QPT GGVLI V G L +D+PP F+Q F L N +GT+ V +
Sbjct 61 SLSFQKITRVITTVDSQPTFDGGVLINVLGRLQCDDDPPHAFSQVFFLKAN-AGTFFVAH 119
Query 139 DCFRLCI 145
D FRL I
Sbjct 120 DIFRLNI 126
> CE06238
Length=133
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/130 (35%), Positives = 71/130 (54%), Gaps = 7/130 (5%)
Query 23 LQLNPHFDSIGPQFVQVYYETFR----NNRA-GLAELYT-ENSLLTYEGTQFRGAAAIVQ 76
+ NP ++S+ F+Q YY F +RA GL++LY ENS +T+EG Q +G I+Q
Sbjct 1 MSFNPDYESVAKAFIQHYYSKFDVGDGMSRAQGLSDLYDPENSYMTFEGQQAKGRDGILQ 60
Query 77 KLQQLP-AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYA 135
K L + + D QP G + ++V G L +++P F+Q F L PN G+Y
Sbjct 61 KFTTLGFTKIQRAITVIDSQPLYDGSIQVMVLGQLKTDEDPINPFSQVFILRPNNQGSYF 120
Query 136 VFNDCFRLCI 145
+ N+ FRL +
Sbjct 121 IGNEIFRLDL 130
> Hs17451119
Length=126
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 46/120 (38%), Positives = 64/120 (53%), Gaps = 3/120 (2%)
Query 27 PHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAV 85
P ++ IG F Q YY+ F N+R L +Y + S LT+E QF+G AA V+KL LP +
Sbjct 5 PVWEPIGSSFNQHYYQLFDNDRTQLGAIYIDASCLTWEVRQFQGKAAAVEKLSSLPFQKI 64
Query 86 SHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRLCI 145
+ L D QPTP ++ +V G L +++P F Q F L G A ND FR +
Sbjct 65 QNSLTAQDHQPTPDSCIIGVVVGQLKADEDPIKGFHQMFLLKNINDGFCA--NDMFRFAL 122
> Hs22048431_2
Length=119
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 63/120 (52%), Gaps = 10/120 (8%)
Query 27 PHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP-AAV 85
P ++ IG F+ YY+ F N+R L +Y + S LT+EG QF+G AAIV+KL L +
Sbjct 5 PIWEQIGSSFIPHYYQLFDNDRPQLGAIYIDASCLTWEGQQFQGKAAIVEKLSSLRFQKI 64
Query 86 SHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRLCI 145
H + D Q TPH ++ +V G L +++ N + T+ ND FRL +
Sbjct 65 QHSITAQDHQLTPHSCIISMVVGQLKADEDRHHG---------NINNTWVCTNDMFRLAL 115
> At1g11570
Length=127
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 57/103 (55%), Gaps = 6/103 (5%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAAVSHQL 89
+ + FV YY F N+R+ L+ LY SLLT+EG G I KL+QLP H L
Sbjct 10 EEVASAFVNHYYHLFDNDRSSLSSLYNPTSLLTFEGQTIYGVDNISNKLKQLPFDQCHHL 69
Query 90 V-TCDCQPTPH----GGVLIIVCGDLAIE-DNPPMKFTQTFNL 126
+ T D QP+ GG+L+ V G + + ++ P++F+Q + L
Sbjct 70 ISTVDSQPSSMAGGCGGILVFVSGSIQLHGEDHPLRFSQVYLL 112
> Hs7019471
Length=140
Score = 64.3 bits (155), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 56/120 (46%), Gaps = 10/120 (8%)
Query 33 GPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAAVSHQLVTC 92
+FV VYY T R L+ LY + L + G G ++ + + LP++ Q+
Sbjct 17 AEEFVNVYYTTMDKRRRLLSRLYMGTATLVWNGNAVSGQESLSEFFEMLPSS-EFQISVV 75
Query 93 DCQP-----TP-HGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVF---NDCFRL 143
DCQP TP VL+++CG + E N F Q F L S + V+ +DCFR
Sbjct 76 DCQPVHDEATPSQTTVLVVICGSVKFEGNKQRDFNQNFILTAQASPSNTVWKIASDCFRF 135
> 7291644
Length=133
Score = 64.3 bits (155), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 57/114 (50%), Gaps = 8/114 (7%)
Query 36 FVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAAVSHQLVTCDCQ 95
F ++YY + N R + LY +N+ L++ G G I Q+LP++ +HQL T D Q
Sbjct 19 FTRLYYASVDNRRQQIGRLYLDNATLSWNGNGAIGRQMIESYFQELPSS-NHQLNTLDAQ 77
Query 96 PTPHGGV------LIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
P V LI+ G + D KF QTF +V + + V +DC+R+
Sbjct 78 PIVDQAVSNQLAYLIMASGSVKFADQQLRKFQQTF-IVTAENDKWKVVSDCYRM 130
> At1g13730
Length=428
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 65/121 (53%), Gaps = 7/121 (5%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQ-----FRGAAAIVQKLQQLPAA 84
++IG FV+ YY + + + + Y ++S+L G+ + AI +++
Sbjct 13 NTIGNSFVEKYYNLLYKSPSQVHQFYLDDSVLGRPGSDGEMVSVKSLKAINEQIMSFDYE 72
Query 85 VSH-QLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
+S Q++T D Q + GV+ +V G L +++ M+F+Q+F LVP +G+Y V ND FR
Sbjct 73 ISKIQILTADSQASYMNGVVTLVTGLLTVKEGQRMRFSQSFFLVP-LNGSYFVLNDVFRY 131
Query 144 C 144
Sbjct 132 V 132
> Hs21361724
Length=197
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 62/148 (41%), Gaps = 16/148 (10%)
Query 10 LEDHLATRS-TERMLQLNPHFDSI---GPQFVQVYYETFRNNRAGLAELYTENSLLTYEG 65
L +H ATRS L + D +FV +YYET R L LY + + L + G
Sbjct 46 LPEHTATRSQMATSLDFKTYVDQACRAAEEFVNIYYETMDKRRRALTRLYLDKATLIWNG 105
Query 66 TQFRGAAAIVQKLQQLPAAVSHQLVTCDCQPT------PHGGVLIIVCGDLAIEDNPPMK 119
G A+ LP++ Q+ DCQP VL++ G + + N
Sbjct 106 NAVSGLDALNNFFDTLPSS-EFQVNMLDCQPVHEQATQSQTTVLVVTSGTVKFDGNKQHF 164
Query 120 FTQTFNL----VPNGSGTYAVFNDCFRL 143
F Q F L PN + + + +DCFR
Sbjct 165 FNQNFLLTAQSTPNNT-VWKIASDCFRF 191
> HsM8923952
Length=142
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 52/121 (42%), Gaps = 12/121 (9%)
Query 33 GPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAAVSHQLVTC 92
+FV +YYET R L LY + + L + G G A+ LP++ Q+
Sbjct 18 AEEFVNIYYETMDKRRRALTRLYLDKATLIWNGNAVSGLDALNNFFDTLPSS-EFQVNML 76
Query 93 DCQPT------PHGGVLIIVCGDLAIEDNPPMKFTQTFNL----VPNGSGTYAVFNDCFR 142
DCQP VL++ G + + N F Q F L PN + + + +DCFR
Sbjct 77 DCQPVHEQATQSQTTVLVVTSGTVKFDGNKQHFFNQNFLLTAQSTPNNT-VWKIASDCFR 135
Query 143 L 143
Sbjct 136 F 136
> CE26780
Length=137
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 65/136 (47%), Gaps = 9/136 (6%)
Query 14 LATRSTERMLQLNPHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAA 73
++ ++T+ + + + + +F+ VYY+ R + LYT+ S + G G +
Sbjct 1 MSMKTTQEINKEDEELCNESKKFMDVYYDVMDRKREKIGFLYTQVSNAVWNGNPINGYDS 60
Query 74 IVQKLQQLPAAVSHQLVTCDCQPTPH-------GGVLIIVCGDLAIEDNPPMKFTQTFNL 126
I + ++ LP+ H + + D Q P GG+L+ V G + ++ + FTQT L
Sbjct 61 ICEFMKALPST-QHDIQSLDAQRLPEGVTGDMSGGMLLNVAGAVTVDGDSKRAFTQTL-L 118
Query 127 VPNGSGTYAVFNDCFR 142
+ G Y V +D FR
Sbjct 119 LGVEDGKYKVKSDRFR 134
> 7299786
Length=690
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 53/118 (44%), Gaps = 2/118 (1%)
Query 27 PHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTY-EGTQFRGAAAIVQKLQQLPAAV 85
P S+G +FV+ YY L Y NS + E G I ++QQL
Sbjct 11 PSPQSVGREFVRQYYTLLNKAPNHLHRFYNHNSSYIHGESKLVVGQREIHNRIQQLNFND 70
Query 86 SHQLVT-CDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFR 142
H ++ D Q T GV++ V G+L+ + P +FTQTF L Y V ND FR
Sbjct 71 CHAKISQVDAQATLGNGVVVQVTGELSNDGQPMRRFTQTFVLAAQSPKKYYVHNDIFR 128
> At2g03640
Length=423
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 13/128 (10%)
Query 25 LNPHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAA 84
++P F +G FVQ YY ++ + + + Y E+S+++ G G ++ L+ +
Sbjct 10 VDPQF--VGNGFVQEYYNHLYDSTSEVHKFYLEDSMISRPG--LDGEIVTIKSLKGINDQ 65
Query 85 VSH--------QLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAV 136
+ +++T D Q T GV+ +V G + D KF+Q+F LV +G+Y V
Sbjct 66 IMSIDYKSSRIEILTADSQSTLKNGVVTLVTGLVIGNDGGRRKFSQSFFLVSR-NGSYFV 124
Query 137 FNDCFRLC 144
ND FR
Sbjct 125 LNDTFRYV 132
> At3g07250_2
Length=666
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 35/117 (29%), Positives = 54/117 (46%), Gaps = 8/117 (6%)
Query 32 IGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQL----PAAV-S 86
+G +F + YY T +N L +LY + S ++ G G + + L P + S
Sbjct 1 VGDEFARQYYNTLQNAPENLYKLYKDKSTISRPG--LDGTMRVFTLSKDLKWRSPGSFDS 58
Query 87 HQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
++ + Q + G+L++V G L + P FTQ F LVP G Y V D FR
Sbjct 59 VKITSVTSQDSLKQGILVVVYGYLTFNERPARHFTQVFFLVPQEKG-YIVCTDMFRF 114
> At5g60980
Length=459
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 38/124 (30%), Positives = 56/124 (45%), Gaps = 7/124 (5%)
Query 26 NPHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLT-----YEGTQFRGAAAIVQKLQQ 80
+P + +G FV+ YY + + Y ++S LT T AI K+
Sbjct 9 SPGAEVVGRAFVEQYYHILHQSPGLVHRFYQDSSFLTRPDVTGAVTTVTTMQAINDKILS 68
Query 81 LP-AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFND 139
L + ++ T D Q + GV+++V G L DN KF+Q+F L P G Y V ND
Sbjct 69 LKYEDYTAEIETADAQESHERGVIVLVTGRLTGNDNVRKKFSQSFFLAPQDKG-YFVLND 127
Query 140 CFRL 143
FR
Sbjct 128 VFRF 131
> At3g25150
Length=486
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 54/123 (43%), Gaps = 11/123 (8%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLP------- 82
D +G FV YY + + Y E S L + G +I LQ +
Sbjct 17 DMVGNAFVPQYYHILHQSPEHVHRFYQEISKLGR--PEENGLMSITSTLQAIDKKIMALG 74
Query 83 -AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCF 141
+S ++ T D Q + GG +++V G L +D+ F+QTF L P +G Y V ND F
Sbjct 75 YGVISAEIATVDTQESHGGGYIVLVTGYLTGKDSVRRTFSQTFFLAPQETG-YFVLNDMF 133
Query 142 RLC 144
R
Sbjct 134 RFI 136
> At5g48650
Length=461
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 53/121 (43%), Gaps = 8/121 (6%)
Query 31 SIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQ-----FRGAAAIVQKLQQLPAAV 85
++G FV YY F N L Y E S + G F I ++L++L
Sbjct 15 TVGSAFVNQYYYIFCNMPEHLPRFYQEISRVGRVGQDGVMRDFSTFQGISEELKRLTYGD 74
Query 86 --SHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRL 143
S ++ + D Q + +GG L+ V G + + KFTQTF L P G + V ND R
Sbjct 75 CNSAEITSYDTQESHNGGFLLFVTGYFTLNERSRRKFTQTFFLAPQEKGFF-VLNDILRF 133
Query 144 C 144
Sbjct 134 V 134
> SPBP8B7.11
Length=434
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/122 (33%), Positives = 54/122 (44%), Gaps = 11/122 (9%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTY--EGTQF---RGAAAIVQKLQQLPAA 84
D IG FVQ YY L YT+ S L + EG G I K+ L
Sbjct 16 DEIGWMFVQEYYTYLNKEPNRLHCFYTKKSTLIHGDEGESISLCHGQQEIHNKILDLDFQ 75
Query 85 VSHQLVT-CDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLV--PNGSGTYAVFNDCF 141
L++ D + +GG++I V G+++ + KF QTF L PNG Y V ND F
Sbjct 76 NCKVLISNVDSLASSNGGIVIQVLGEMSNKGKLSRKFAQTFFLAEQPNG---YFVLNDIF 132
Query 142 RL 143
R
Sbjct 133 RF 134
> YNR051c_1
Length=207
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 56/130 (43%), Gaps = 21/130 (16%)
Query 36 FVQVYYETFRNNRAGLAELYTENSLLTYEGTQFR---------------GAAAIVQKLQQ 80
F+Q YYE R + + LA Y + LT+ Q + G I + +
Sbjct 12 FLQNYYERMRTDPSKLAYFYASTAELTHTNYQSKSTNEKDDVLPTVKVTGRENINKFFSR 71
Query 81 LPAAVSH---QLVTCDCQPTPH--GGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSG-TY 134
A V +L T D Q T H +LI+ G++ P KF QTF L+P+ +G T+
Sbjct 72 NDAKVRSLKLKLDTIDFQYTGHLHKSILIMATGEMFWTGTPVYKFCQTFILLPSSNGSTF 131
Query 135 AVFNDCFRLC 144
+ ND R
Sbjct 132 DITNDIIRFI 141
> CE06150
Length=537
Score = 44.7 bits (104), Expect = 6e-05, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 48/114 (42%), Gaps = 6/114 (5%)
Query 30 DSIGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAAIVQKLQQLPAAVSHQL 89
+ +G F +Y T NR + + Y S + G+ IV+ LP ++
Sbjct 98 EQVGGAFCHQFYITVSENRKAITKFYGHESKFYLDDQVVTGSQEIVKLYNHLPETTHFKI 157
Query 90 VTCDCQPTPHG-GVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFR 142
+ PTPH GV+I V G + + F Q+F L G Y V +D F+
Sbjct 158 QSIKGYPTPHKQGVIINVIGTVNLR-----PFLQSFLLGQQGQKKYYVESDAFQ 206
> Hs19923399
Length=482
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 37/122 (30%), Positives = 49/122 (40%), Gaps = 11/122 (9%)
Query 32 IGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRGAAA--------IVQKLQQLPA 83
+G +FV+ YY L Y NS + G G I K+ L
Sbjct 11 VGREFVRQYYTLLNKAPEYLHRFYGRNSSYVHGGVDASGKPQEAVYGQNDIHHKVLSLNF 70
Query 84 AVSH-QLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGS--GTYAVFNDC 140
+ H ++ D T GV++ V G L+ P KF QTF L P GS + V ND
Sbjct 71 SECHTKIRHVDAHATLSDGVVVQVMGLLSNSGQPERKFMQTFVLAPEGSVPNKFYVHNDM 130
Query 141 FR 142
FR
Sbjct 131 FR 132
> Hs5031703
Length=466
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 35/122 (28%), Positives = 51/122 (41%), Gaps = 11/122 (9%)
Query 32 IGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQFRG--AAAIVQKLQQLPAAVSHQL 89
+G +FV+ YY L Y +NS + G G A A+ + + +S
Sbjct 11 VGREFVRQYYTLLNQAPDMLHRFYGKNSSYVHGGLDSNGKPADAVYGQKEIHRKVMSQNF 70
Query 90 VTC-------DCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGS--GTYAVFNDC 140
C D T + GV++ V G L+ + +F QTF L P GS + V ND
Sbjct 71 TNCHTKIRHVDAHATLNDGVVVQVMGLLSNNNQALRRFMQTFVLAPEGSVANKFYVHNDI 130
Query 141 FR 142
FR
Sbjct 131 FR 132
> SPAPB1A10.03
Length=200
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 12/93 (12%)
Query 35 QFVQVYYETFRNNRAGLAELYTENSLLTYEGT--QFRGAAAIVQKLQQLPAAV----SHQ 88
+FVQ YY + NR G+AE Y ENSL+ + G Q +++ L V S Q
Sbjct 10 EFVQRYYSSLDTNRNGIAEFYRENSLILWNGKPMQVTEFTSMIVNLPYSKTKVEDFDSQQ 69
Query 89 LVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFT 121
++ D ++I+V G + + P F+
Sbjct 70 VMGNDM------NIIIVVSGTIRFDGKKPHVFS 96
> At3g55540
Length=334
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 55/117 (47%), Gaps = 12/117 (10%)
Query 36 FVQVYYETFRNNRAGLAELYTENSLLTYEGT-------QFRGAAAIVQKLQ-QLPAAVSH 87
FV+VYYE GL +Y S+++ + + G AI +++ + A +
Sbjct 170 FVKVYYELPMREELGL--MYVNESIMSRPTSTSGRTMVEMPGLDAINKRVSNEHKRASNF 227
Query 88 QLVTCDCQPTP--HGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFR 142
L + D Q + + I+VCG + ++D KF Q F + +G+Y ++ND R
Sbjct 228 ILNSVDYQISRSFKDRMFIMVCGFVTLDDKTERKFLQFFYVTRCHNGSYVIYNDILR 284
> At3g07240
Length=369
Score = 35.0 bits (79), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 48/119 (40%), Gaps = 7/119 (5%)
Query 32 IGPQFVQVYYETFRNNRAGLAELYTENSLLTYEGTQ--FRGAA--AIVQKLQQL-PAAV- 85
IG F + YY +N+ L Y + S +T G R + I++ L L P
Sbjct 19 IGDGFAEHYYNNLQNSPEILPGYYKDVSKITRPGLDGTMRSSTLPDIIEDLDMLSPGGFD 78
Query 86 SHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDCFRLC 144
S ++ + Q + G+ + V G + P FTQ F P G + V D F+
Sbjct 79 SVEVTSVMSQDSHDKGIRVAVDGYFTFNERPARNFTQNFTFAPQEKGLF-VSTDMFKFV 136
> At1g69250
Length=427
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 50/122 (40%), Gaps = 8/122 (6%)
Query 27 PHFDSIGPQFVQVYYETFRNNRAGLAELYTENSLLT---YEGT--QFRGAAAIVQKLQQL 81
P I +FV+ YY LY + S+++ GT F AI + +
Sbjct 8 PSAQDIAAEFVRQYYHVLGQLPHEARRLYVDASVVSRPDVTGTMMSFTSVEAINKHILSC 67
Query 82 P-AAVSHQLVTCDCQPTPHGGVLIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDC 140
++++ D Q + G+ I+V G + +DN KF+Q F L T V ND
Sbjct 68 DFENTKFEVLSVDSQNSLEDGIFIMVIGFMTGKDNQRRKFSQMFYLA--RQNTLVVLNDM 125
Query 141 FR 142
R
Sbjct 126 LR 127
> At5g40440
Length=520
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 29/68 (42%), Gaps = 9/68 (13%)
Query 54 LYTENSLLTYEGTQFRGAAAIVQKLQQ--------LPA-AVSHQLVTCDCQPTPHGGVLI 104
LYTE S+ ++ G G+ I L LP+ + H + C+P GGV+I
Sbjct 391 LYTETSVFSFSGKHNTGSTEIFSALSDIRNTLTGDLPSEKLVHVVEKLHCKPCGSGGVII 450
Query 105 IVCGDLAI 112
G +
Sbjct 451 RAVGSFIV 458
> Hs19923308
Length=318
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 22/42 (52%), Gaps = 10/42 (23%)
Query 82 PAAVSHQLVTC----------DCQPTPHGGVLIIVCGDLAIE 113
PA S+ LVTC D Q TP G+LIIV G + +E
Sbjct 174 PAGHSYILVTCLGLSYDGLLGDDQSTPKTGLLIIVLGMILME 215
> YCR019w
Length=363
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 103 LIIVCGDLAIEDNPPMKFTQTFNLVPNGSGTYAVFNDC 140
L+I G I+D K+ + VP G GT+A+ C
Sbjct 13 LVITNGMFIIDDIERSKYNIHYKNVPGGGGTFAILGAC 50
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40