bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0905_orf1
Length=403
Score E
Sequences producing significant alignments: (Bits) Value
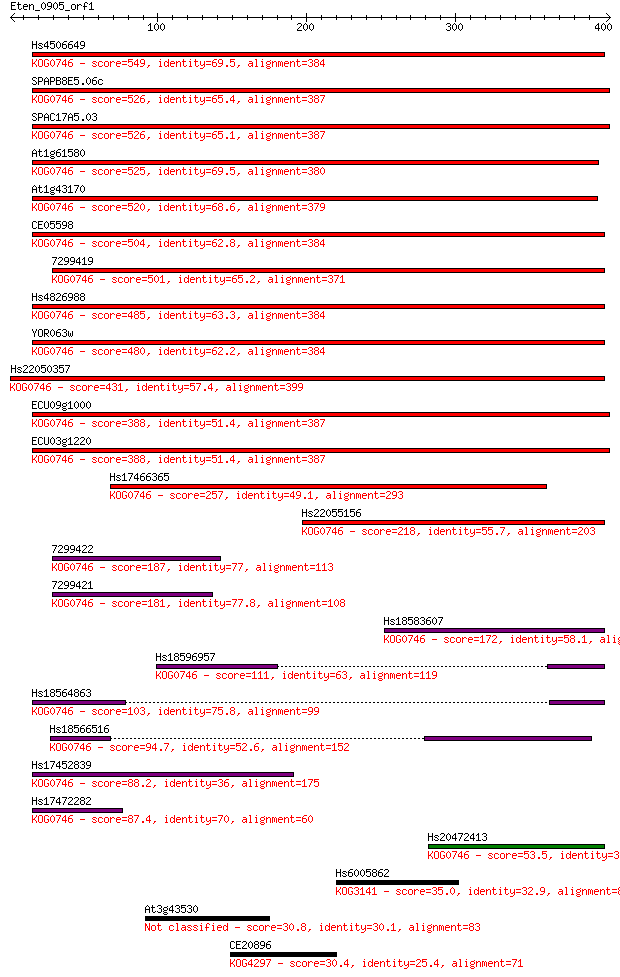
Hs4506649 549 5e-156
SPAPB8E5.06c 526 2e-149
SPAC17A5.03 526 4e-149
At1g61580 525 5e-149
At1g43170 520 2e-147
CE05598 504 2e-142
7299419 501 1e-141
Hs4826988 485 8e-137
YOR063w 480 3e-135
Hs22050357 431 2e-120
ECU09g1000 388 1e-107
ECU03g1220 388 1e-107
Hs17466365 257 4e-68
Hs22055156 218 2e-56
7299422 187 3e-47
7299421 181 3e-45
Hs18583607 172 9e-43
Hs18596957 111 3e-24
Hs18564863 103 5e-22
Hs18566516 94.7 3e-19
Hs17452839 88.2 3e-17
Hs17472282 87.4 5e-17
Hs20472413 53.5 7e-07
Hs6005862 35.0 0.29
At3g43530 30.8 5.5
CE20896 30.4 6.7
> Hs4506649
Length=403
Score = 549 bits (1414), Expect = 5e-156, Method: Compositional matrix adjust.
Identities = 267/394 (67%), Positives = 319/394 (80%), Gaps = 10/394 (2%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PRHGSLGF PRKR RHRGKVK+FPKDDPSKP HLTAF+GYKAGMTHIVREV
Sbjct 1 MSHRKFSAPRHGSLGFLPRKRSSRHRGKVKSFPKDDPSKPVHLTAFLGYKAGMTHIVREV 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
+RPGSK++KKEVVEAVTIVE+PPMV VG+VGY+ETP+GLR TV+A H+S+EC+RRFYK
Sbjct 61 DRPGSKVNKKEVVEAVTIVETPPMVVVGIVGYVETPRGLRTFKTVFAEHISDECKRRFYK 120
Query 136 NWYKSKKKAFTRYSKKYA-ENNKMQAEID--SIKQHCAVIRAICHTQPSKTPTGLRKAHI 192
NW+KSKKKAFT+Y KK+ E+ K Q E D S+K++C VIR I HTQ P +KAH+
Sbjct 121 NWHKSKKKAFTKYCKKWQDEDGKKQLEKDFSSMKKYCQVIRVIAHTQMRLLPLRQKKAHL 180
Query 193 MEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRL 252
MEIQVNGGTVA+K+D+ + E VP++ VF QDEM+DVIGVTKG G KGV SR+ +L
Sbjct 181 MEIQVNGGTVAEKLDWARERLEQQVPVNQVFGQDEMIDVIGVTKGKGYKGVTSRWHTKKL 240
Query 253 PRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNG---- 308
PRKTHRGLRKVACIGAWHPARV F V R GQKGY HRTE+NKK+Y+IG G ++G
Sbjct 241 PRKTHRGLRKVACIGAWHPARVAFSVARAGQKGYHHRTEINKKIYKIGQGYLIKDGKLIK 300
Query 309 ---STDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRAL 365
STD DL++K I P+GGF HYGEV +DF+M+KGC+VG KKR +T+RK+L+ +T RRAL
Sbjct 301 NNASTDYDLSDKSINPLGGFVHYGEVTNDFVMLKGCVVGTKKRVLTLRKSLLVQTKRRAL 360
Query 366 ENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
E I LKFIDT+SK+GHGRFQT +EK F GPLK+
Sbjct 361 EKIDLKFIDTTSKFGHGRFQTMEEKKAFMGPLKK 394
> SPAPB8E5.06c
Length=388
Score = 526 bits (1356), Expect = 2e-149, Method: Compositional matrix adjust.
Identities = 253/388 (65%), Positives = 316/388 (81%), Gaps = 1/388 (0%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSH KFE+PRHGSLGF PRKR R RGKVKAFPKDD SKP HLTAF+GYKAGMTHIVR++
Sbjct 1 MSHCKFEQPRHGSLGFLPRKRASRQRGKVKAFPKDDASKPVHLTAFLGYKAGMTHIVRDL 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
+RPGSK+HK+E++EAVTI+E+PPMV VG+VGY+ETP+GLR+L+TVWA HLSEE +RRFYK
Sbjct 61 DRPGSKMHKREILEAVTIIETPPMVVVGVVGYVETPRGLRSLTTVWAEHLSEEVKRRFYK 120
Query 136 NWYKSKKKAFTRYSKKYAENNK-MQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHIME 194
NW+KSKKKAFT+Y+KKYAE+ + + E++ IK++C+V+R + HTQ KTP +KAH+ME
Sbjct 121 NWFKSKKKAFTKYAKKYAESTQSINRELERIKKYCSVVRVLAHTQIRKTPLAQKKAHLME 180
Query 195 IQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLPR 254
IQVNGG+VADKV++ + FE V I + FEQ+EM+DVIGVT+G G +G +R+G RLPR
Sbjct 181 IQVNGGSVADKVEWAREHFEKTVDIKSTFEQNEMIDVIGVTRGKGNEGTTARWGTKRLPR 240
Query 255 KTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNGSTDADL 314
KTHRGLRKVACIGAWHPA VQ+ V R G GY HRT++N K+YRIG+G D +N STD D
Sbjct 241 KTHRGLRKVACIGAWHPANVQWTVARAGNAGYMHRTQLNSKIYRIGAGDDAKNASTDFDA 300
Query 315 TEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFID 374
TEKRITPMGGF YG V +DF+M+ G G KR +T+RK+L+ TSR+ALE + LK+ID
Sbjct 301 TEKRITPMGGFVRYGVVENDFVMLNGATPGPVKRVLTLRKSLLTHTSRKALEPVSLKWID 360
Query 375 TSSKWGHGRFQTSDEKAKFYGPLKRTAA 402
T+SK+GHGRFQT E +F G LK+ A
Sbjct 361 TASKFGHGRFQTPAEAKQFLGTLKKDVA 388
> SPAC17A5.03
Length=388
Score = 526 bits (1354), Expect = 4e-149, Method: Compositional matrix adjust.
Identities = 252/388 (64%), Positives = 316/388 (81%), Gaps = 1/388 (0%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSH KFE+PRHGSLGF PRKR R RGKVKAFPKDD SKP HLTAF+GYKAGMTHIVR++
Sbjct 1 MSHCKFEQPRHGSLGFLPRKRASRQRGKVKAFPKDDASKPVHLTAFLGYKAGMTHIVRDL 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
+RPGSK+HK+E++EAVT++E+PPMV VG+VGY+ETP+GLR+L+TVWA HLSEE +RRFYK
Sbjct 61 DRPGSKMHKREILEAVTVIETPPMVVVGVVGYVETPRGLRSLTTVWAEHLSEEVKRRFYK 120
Query 136 NWYKSKKKAFTRYSKKYAENNK-MQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHIME 194
NW+KSKKKAFT+Y+KKYAE+ + + E++ IK++C+V+R + HTQ KTP +KAH+ME
Sbjct 121 NWFKSKKKAFTKYAKKYAESTQSINRELERIKKYCSVVRVLAHTQIRKTPLAQKKAHLME 180
Query 195 IQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLPR 254
IQVNGG+VADKV++ + FE V I + FEQ+EM+DVIGVT+G G +G +R+G RLPR
Sbjct 181 IQVNGGSVADKVEWAREHFEKTVDIKSTFEQNEMIDVIGVTRGKGNEGTTARWGTKRLPR 240
Query 255 KTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNGSTDADL 314
KTHRGLRKVACIGAWHPA VQ+ V R G GY HRT++N K+YRIG+G D +N STD D
Sbjct 241 KTHRGLRKVACIGAWHPANVQWTVARAGNAGYMHRTQLNSKIYRIGAGDDAKNASTDFDA 300
Query 315 TEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFID 374
TEKRITPMGGF YG V +DF+M+ G G KR +T+RK+L+ TSR+ALE + LK+ID
Sbjct 301 TEKRITPMGGFVRYGVVENDFVMLNGATPGPVKRVLTLRKSLLTHTSRKALEPVSLKWID 360
Query 375 TSSKWGHGRFQTSDEKAKFYGPLKRTAA 402
T+SK+GHGRFQT E +F G LK+ A
Sbjct 361 TASKFGHGRFQTPAEAKQFLGTLKKDVA 388
> At1g61580
Length=390
Score = 525 bits (1353), Expect = 5e-149, Method: Compositional matrix adjust.
Identities = 264/386 (68%), Positives = 317/386 (82%), Gaps = 7/386 (1%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKFE PRHGSLGF PRKR RHRGKVKAFPKDDP+KP LT+F+GYKAGMTHIVR+V
Sbjct 1 MSHRKFEHPRHGSLGFLPRKRASRHRGKVKAFPKDDPTKPCRLTSFLGYKAGMTHIVRDV 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
E+PGSKLHKKE EAVTI+E+PPMV VG+VGY++TP+GLR+L TVWA HLSEE RRRFYK
Sbjct 61 EKPGSKLHKKETCEAVTIIETPPMVVVGVVGYVKTPRGLRSLCTVWAQHLSEELRRRFYK 120
Query 136 NWYKSKKKAFTRYSKKY-AENNK--MQAEIDSIKQHCAVIRAICHTQPSKTPTGL--RKA 190
NW KSKKKAFTRYSKK+ E K +Q++++ +K++C+VIR + HTQ K GL +KA
Sbjct 121 NWAKSKKKAFTRYSKKHETEEGKKDIQSQLEKMKKYCSVIRVLAHTQIRKMK-GLKQKKA 179
Query 191 HIMEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVT 250
H+ EIQ+NGG +A KVD+ +FE VP+ A+F++DEM+D+IGVTKG G +GVV+R+GVT
Sbjct 180 HLNEIQINGGDIAKKVDYACSLFEKQVPVDAIFQKDEMIDIIGVTKGKGYEGVVTRWGVT 239
Query 251 RLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGS-GSDPRNGS 309
RLPRKTHRGLRKVACIGAWHPARV + V R GQ GY HRTEMNKKVYR+G G + +
Sbjct 240 RLPRKTHRGLRKVACIGAWHPARVSYTVARAGQNGYHHRTEMNKKVYRVGKVGQETHSAM 299
Query 310 TDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIH 369
T+ D TEK ITPMGGFPHYG V+ D++MIKGC VG KKR +T+R+TL+ +TSR A+E I
Sbjct 300 TEYDRTEKDITPMGGFPHYGIVKEDYLMIKGCCVGPKKRVVTLRQTLLKQTSRLAMEEIK 359
Query 370 LKFIDTSSKWGHGRFQTSDEKAKFYG 395
LKFID +S GHGRFQTS EKAKFYG
Sbjct 360 LKFIDAASNGGHGRFQTSQEKAKFYG 385
> At1g43170
Length=389
Score = 520 bits (1339), Expect = 2e-147, Method: Compositional matrix adjust.
Identities = 260/385 (67%), Positives = 314/385 (81%), Gaps = 7/385 (1%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKFE PRHGSLGF PRKR RHRGKVKAFPKDD +KP TAFMGYKAGMTHIVREV
Sbjct 1 MSHRKFEHPRHGSLGFLPRKRANRHRGKVKAFPKDDQTKPCKFTAFMGYKAGMTHIVREV 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
E+PGSKLHKKE EAVTI+E+P MV VG+V Y++TP+GLR+L+TVWA HLSEE RRRFYK
Sbjct 61 EKPGSKLHKKETCEAVTIIETPAMVVVGVVAYVKTPRGLRSLNTVWAQHLSEEVRRRFYK 120
Query 136 NWYKSKKKAFTRYSKKY-AENNK--MQAEIDSIKQHCAVIRAICHTQPSKTPTGL--RKA 190
NW KSKKKAFT Y+K+Y +E+ K +QA+++ +K++ VIR + HTQ K GL +KA
Sbjct 121 NWAKSKKKAFTGYAKQYDSEDGKKGIQAQLEKMKKYATVIRVLAHTQIRKMK-GLKQKKA 179
Query 191 HIMEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVT 250
H+MEIQ+NGGT+A KVDF FE +PI AVF++DEM+D+IGVTKG G +GVV+R+GVT
Sbjct 180 HMMEIQINGGTIAQKVDFAYSFFEKQIPIEAVFQKDEMIDIIGVTKGKGYEGVVTRWGVT 239
Query 251 RLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGS-GSDPRNGS 309
RLPRKTHRGLRKVACIGAWHPARV + V R GQ GY HRTE+NKK+YR+G G++
Sbjct 240 RLPRKTHRGLRKVACIGAWHPARVSYTVARAGQNGYHHRTELNKKIYRLGKVGTEAHTAM 299
Query 310 TDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIH 369
T+ D TEK +TPMGGFPHYG V+ D++MIKGC VG KKR +T+R++L+ +TSR ALE I
Sbjct 300 TEYDRTEKDVTPMGGFPHYGIVKDDYLMIKGCCVGPKKRVVTLRQSLLTQTSRLALEEIK 359
Query 370 LKFIDTSSKWGHGRFQTSDEKAKFY 394
LKFIDT+S +GHGRFQTS EK +FY
Sbjct 360 LKFIDTASIFGHGRFQTSLEKMRFY 384
> CE05598
Length=401
Score = 504 bits (1297), Expect = 2e-142, Method: Compositional matrix adjust.
Identities = 241/391 (61%), Positives = 301/391 (76%), Gaps = 7/391 (1%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PRHG +GF P+KR R +RG++KAFPKDD SKP HLTAF+GYKAGMTHIVR+V
Sbjct 1 MSHRKFSAPRHGHMGFTPKKRSRTYRGRIKAFPKDDKSKPIHLTAFLGYKAGMTHIVRDV 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
++PGSK++KKEVVEAVTIVE+PPMV G+ GY++TPQG RAL+T+WA HLSEE RRRFY
Sbjct 61 DKPGSKVNKKEVVEAVTIVETPPMVIAGVTGYVDTPQGPRALTTIWAEHLSEEARRRFYS 120
Query 136 NWYKSKKKAFTRYSKKYAENNK---MQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHI 192
NW KSKKKAFT+Y+KK+ + + ++A+ +K++C+ IR I HTQ +KAH+
Sbjct 121 NWAKSKKKAFTKYAKKWQDEDGKKLIEADFAKLKKYCSSIRVIAHTQMKILRRRQKKAHL 180
Query 193 MEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRL 252
+EIQVNGGT+ KVD+ + E V + VF QDEM+D IGVT+GHG KGV SR+ +L
Sbjct 181 VEIQVNGGTIEQKVDWAREHLEKQVQVDTVFAQDEMIDTIGVTRGHGFKGVTSRWHTKKL 240
Query 253 PRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGS----DPRNG 308
PRKTH+GLRKVACIGAWHP+RV F V R GQKG+ HRT +N K+YRIG + NG
Sbjct 241 PRKTHKGLRKVACIGAWHPSRVAFTVARAGQKGFHHRTIINNKIYRIGKSALTEEGKNNG 300
Query 309 STDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENI 368
ST+ DLT+K ITPMGGFP YG V D+IM++G ++G KKR IT+RK+L+ +T R A E I
Sbjct 301 STEFDLTQKTITPMGGFPRYGIVNQDYIMLRGAVLGPKKRLITLRKSLITQTKRVAHEKI 360
Query 369 HLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
+LK+IDTSSK GHGRFQT+ EK F G LKR
Sbjct 361 NLKWIDTSSKTGHGRFQTTAEKRAFMGKLKR 391
> 7299419
Length=403
Score = 501 bits (1290), Expect = 1e-141, Method: Compositional matrix adjust.
Identities = 242/381 (63%), Positives = 293/381 (76%), Gaps = 10/381 (2%)
Query 29 LGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREVERPGSKLHKKEVV 88
+ F P+KR RHRGKVKAFPKDD SKP HLT F+GYKAGMTHIVRE +RPGSK++KKEVV
Sbjct 1 MAFYPKKRSARHRGKVKAFPKDDASKPVHLTCFIGYKAGMTHIVREADRPGSKINKKEVV 60
Query 89 EAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYKNWYKSKKKAFTRY 148
EAVT++E+PPM+ VG VGYIETP GLRAL VWA HLSEECRRRFYKNWYKSKKKAFT+
Sbjct 61 EAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSEECRRRFYKNWYKSKKKAFTKA 120
Query 149 SKKYAEN---NKMQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHIMEIQVNGGTVADK 205
SKK+ ++ ++ + + ++C VIR I H+Q +KAH+MEIQ+NGG++ DK
Sbjct 121 SKKWTDDLGKKSIENDFRKMLRYCKVIRVIAHSQIRLIKQRQKKAHVMEIQLNGGSIEDK 180
Query 206 VDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLPRKTHRGLRKVAC 265
V + + E P+ +S VF QDEM+D +GVTKG G KGV SR+ +LPRKTH+GLRKVAC
Sbjct 181 VKWAREHLEKPIQVSNVFGQDEMIDCVGVTKGKGFKGVTSRWHTKKLPRKTHKGLRKVAC 240
Query 266 IGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNG-------STDADLTEKR 318
IGAWHP+RV V R GQKGY HRTE+NKK+YRIG+G ++G ST+ DLT+K
Sbjct 241 IGAWHPSRVSTTVARAGQKGYHHRTEINKKIYRIGAGIHTKDGKVIKNNASTEYDLTDKS 300
Query 319 ITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFIDTSSK 378
ITPMGGFPHYGEV +DF+MIKGC +G KKR IT+RK+L+ T R ALE I LKFIDTSSK
Sbjct 301 ITPMGGFPHYGEVNNDFVMIKGCCIGSKKRIITLRKSLLKHTKRSALEQIKLKFIDTSSK 360
Query 379 WGHGRFQTSDEKAKFYGPLKR 399
GHGRFQT +K F GPLK+
Sbjct 361 MGHGRFQTPADKLAFMGPLKK 381
> Hs4826988
Length=407
Score = 485 bits (1248), Expect = 8e-137, Method: Compositional matrix adjust.
Identities = 243/394 (61%), Positives = 302/394 (76%), Gaps = 10/394 (2%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PRHG LGF P KR RHRGKVK +P+DDPS+P HLTAF+GYKAGMTH +REV
Sbjct 1 MSHRKFSAPRHGHLGFLPHKRSHRHRGKVKTWPRDDPSQPVHLTAFLGYKAGMTHTLREV 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
RPG K+ K+E VEAVTIVE+PP+V VG+VGY+ TP+GLR+ T++A HLS+ECRRRFYK
Sbjct 61 HRPGLKISKREEVEAVTIVETPPLVVVGVVGYVATPRGLRSFKTIFAEHLSDECRRRFYK 120
Query 136 NWYKSKKKAFTRYSKKYAENN---KMQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHI 192
+W+KSKKKAFT+ K++ + + ++Q + ++K++C VIR I HTQ P +KAHI
Sbjct 121 DWHKSKKKAFTKACKRWRDTDGKKQLQKDFAAMKKYCKVIRVIVHTQMKLLPFRQKKAHI 180
Query 193 MEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRL 252
MEIQ+NGGTVA+KV + E VP+ +VF Q E++DVI VTKG GVKGV SR+ +L
Sbjct 181 MEIQLNGGTVAEKVAWAQARLEKQVPVHSVFSQSEVIDVIAVTKGRGVKGVTSRWHTKKL 240
Query 253 PRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSD-------P 305
PRKTH+GLRKVACIGAWHPARV + R GQKGY HRTE+NKK++RIG G
Sbjct 241 PRKTHKGLRKVACIGAWHPARVGCSIARAGQKGYHHRTELNKKIFRIGRGPHMEDGKLVK 300
Query 306 RNGSTDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRAL 365
N ST D+T K ITP+GGFPHYGEV +DF+M+KGCI G KKR IT+RK+L+ SR+A+
Sbjct 301 NNASTSYDVTAKSITPLGGFPHYGEVNNDFVMLKGCIAGTKKRVITLRKSLLVHHSRQAV 360
Query 366 ENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
ENI LKFIDT+SK+GHGRFQT+ EK F GP K+
Sbjct 361 ENIELKFIDTTSKFGHGRFQTAQEKRAFMGPQKK 394
> YOR063w
Length=387
Score = 480 bits (1235), Expect = 3e-135, Method: Compositional matrix adjust.
Identities = 239/385 (62%), Positives = 306/385 (79%), Gaps = 1/385 (0%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRK+E PRHG LGF PRKR R +VKAFPKDD SKP LT+F+GYKAGMT IVR++
Sbjct 1 MSHRKYEAPRHGHLGFLPRKRAASIRARVKAFPKDDRSKPVALTSFLGYKAGMTTIVRDL 60
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
+RPGSK HK+EVVEAVT+V++PP+V VG+VGY+ETP+GLR+L+TVWA HLS+E +RRFYK
Sbjct 61 DRPGSKFHKREVVEAVTVVDTPPVVVVGVVGYVETPRGLRSLTTVWAEHLSDEVKRRFYK 120
Query 136 NWYKSKKKAFTRYSKKYAENNK-MQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHIME 194
NWYKSKKKAFT+YS KYA++ ++ E+ IK++ +V+R + HTQ KTP +KAH+ E
Sbjct 121 NWYKSKKKAFTKYSAKYAQDGAGIERELARIKKYASVVRVLVHTQIRKTPLAQKKAHLAE 180
Query 195 IQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLPR 254
IQ+NGG++++KVD+ + FE V + +VFEQ+EM+D I VTKGHG +GV R+G +LPR
Sbjct 181 IQLNGGSISEKVDWAREHFEKTVAVDSVFEQNEMIDAIAVTKGHGFEGVTHRWGTKKLPR 240
Query 255 KTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNGSTDADL 314
KTHRGLRKVACIGAWHPA V + V R GQ+GY RT +N K+YR+G G D NG+T D
Sbjct 241 KTHRGLRKVACIGAWHPAHVMWSVARAGQRGYHSRTSINHKIYRVGKGDDEANGATSFDR 300
Query 315 TEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFID 374
T+K ITPMGGF HYGE+++DFIM+KGCI G +KR +T+RK+L TSR+ALE + LK+ID
Sbjct 301 TKKTITPMGGFVHYGEIKNDFIMVKGCIPGNRKRIVTLRKSLYTNTSRKALEEVSLKWID 360
Query 375 TSSKWGHGRFQTSDEKAKFYGPLKR 399
T+SK+G GRFQT EK F G LK+
Sbjct 361 TASKFGKGRFQTPAEKHAFMGTLKK 385
> Hs22050357
Length=432
Score = 431 bits (1107), Expect = 2e-120, Method: Compositional matrix adjust.
Identities = 229/409 (55%), Positives = 289/409 (70%), Gaps = 16/409 (3%)
Query 1 LDVEFCRILFHFYSKMSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTA 60
L + F + F MSHR+F PRHGSLGF P++R R+H GKVK+FPKDDPSK HLTA
Sbjct 21 LKIPFFALPAGFDGMMSHRRFSAPRHGSLGFLPQQRSRKHHGKVKSFPKDDPSKLVHLTA 80
Query 61 FMGYKAGMTHIVREVERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTV 120
F+GYKAGMT+I+REV+RPGSK++KKEV+EAVTIVE PPMV V +VG++ETP+GL+ +TV
Sbjct 81 FLGYKAGMTNIMREVDRPGSKVNKKEVIEAVTIVEIPPMVVVAIVGFVETPRGLQTFNTV 140
Query 121 WAGHLSEECRRRFYKNWYKSKKKAFTRYSKK-YAENNKMQAEID--SIKQHCAVIRAICH 177
+A H+S+ C+R FYKN +KSKKKAFT+Y KK E+ K Q E D S+K++ VI I +
Sbjct 141 FAEHISDACKRHFYKNRHKSKKKAFTKYCKKCQDEDGKKQLEKDFSSMKKYRQVICIIAY 200
Query 178 TQPSKTPTGLRKAHIMEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKG 237
TQ P RKAH+MEIQV GGT+A K+D+ + E VP++ VFEQD+M+DVI +TKG
Sbjct 201 TQMCLLPLHQRKAHLMEIQVKGGTMAKKLDWAHERLEQQVPVNQVFEQDKMIDVIRLTKG 260
Query 238 HGVKGVVSRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVY 297
+G KGV SR+ +LP KTHRGL KVAC G HPA V V QKGY H TE+N K+Y
Sbjct 261 NGYKGVTSRWHTKKLPHKTHRGLCKVACTGTQHPAPVALPVACIWQKGYCHSTEIN-KIY 319
Query 298 RIGSGSDPR-------NGSTDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPI 350
+I G R N STD DL P+G F HYGEV +DF+M+K C+V KK+ +
Sbjct 320 KISQGYFVRDSKLIKNNASTDYDL-----HPLGAFVHYGEVTNDFVMLKDCVVDTKKQVL 374
Query 351 TMRKTLVPRTSRRALENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
T+ K+L T RR LE I LKFIDT+SK+GHG FQT +EK F PLK+
Sbjct 375 TLHKSLPVPTKRRPLEKIDLKFIDTTSKFGHGCFQTMEEKTAFMAPLKK 423
> ECU09g1000
Length=383
Score = 388 bits (996), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 199/389 (51%), Positives = 266/389 (68%), Gaps = 12/389 (3%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVR-E 74
MS RKF PRHGSL FCPRKR + R AFP DD S+P HLT FM YKAGMTH+VR +
Sbjct 1 MSCRKFNAPRHGSLQFCPRKRSKTIRPAAGAFPADDRSQPVHLTGFMAYKAGMTHVVRTK 60
Query 75 VERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFY 134
+ +K +E+++AVT++E+PPMV G+VGY +T GLR L V A ++S+ RR +
Sbjct 61 TQVAKNKQLSREIMDAVTVLEAPPMVVYGIVGYEKTVTGLRRLPIVTAAYVSDGVLRRMF 120
Query 135 KNWYKSKKKAFTRYSKKYAENNKMQAEIDSIKQHCAVIRAICHTQPSKTP-TGLRKAHIM 193
N Y SK+ A ++ + + ++ ++ IK+ +R + TQP+ GL+KAHI
Sbjct 121 GNRYASKESA-----GQFCKGSVCESRVEMIKERAHCVRVLVQTQPTLIKGLGLKKAHIA 175
Query 194 EIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLP 253
EIQVNGG++++KV++ E + I VF +E +D IGVTKG G +G V RFGV + P
Sbjct 176 EIQVNGGSISEKVEWALGRLEKEIAIGEVFGVNENIDTIGVTKGKGFQGTVKRFGVRKQP 235
Query 254 RKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNGSTDAD 313
RK+ +G+RKVACIGAWHP+RV + + R GQ G+ RTE NK+VY IG+GS N T+ D
Sbjct 236 RKSRKGIRKVACIGAWHPSRVMYSIARAGQMGFHRRTEKNKRVYMIGNGSS--NIKTEFD 293
Query 314 LTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFI 373
LTEK I+PMGGFPHYGEVR+DFIM+KG +VG +KR +T+RK+L+ +RA E + +KF+
Sbjct 294 LTEKPISPMGGFPHYGEVRNDFIMVKGAVVGARKRVVTLRKSLL---RQRAGEELVIKFV 350
Query 374 DTSSKWGHGRFQTSDEKAKFYGPLKRTAA 402
DTSSK GHGRFQTS EK FYG K A
Sbjct 351 DTSSKIGHGRFQTSAEKKAFYGARKADIA 379
> ECU03g1220
Length=383
Score = 388 bits (996), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 199/389 (51%), Positives = 266/389 (68%), Gaps = 12/389 (3%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVR-E 74
MS RKF PRHGSL FCPRKR + R AFP DD S+P HLT FM YKAGMTH+VR +
Sbjct 1 MSCRKFNAPRHGSLQFCPRKRSKTIRPAAGAFPADDRSQPVHLTGFMAYKAGMTHVVRTK 60
Query 75 VERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFY 134
+ +K +E+++AVT++E+PPMV G+VGY +T GLR L V A ++S+ RR +
Sbjct 61 TQVAKNKQLSREIMDAVTVLEAPPMVVYGIVGYEKTVTGLRRLPIVTAAYVSDGVLRRMF 120
Query 135 KNWYKSKKKAFTRYSKKYAENNKMQAEIDSIKQHCAVIRAICHTQPSKTP-TGLRKAHIM 193
N Y SK+ A ++ + + ++ ++ IK+ +R + TQP+ GL+KAHI
Sbjct 121 GNRYASKESA-----GQFCKGSVCESRVEMIKERAHCVRVLVQTQPTLIKGLGLKKAHIA 175
Query 194 EIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLP 253
EIQVNGG++++KV++ E + I VF +E +D IGVTKG G +G V RFGV + P
Sbjct 176 EIQVNGGSISEKVEWALGRLEKEIAIGEVFGVNENIDTIGVTKGKGFQGTVKRFGVRKQP 235
Query 254 RKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNGSTDAD 313
RK+ +G+RKVACIGAWHP+RV + + R GQ G+ RTE NK+VY IG+GS N T+ D
Sbjct 236 RKSRKGIRKVACIGAWHPSRVMYSIARAGQMGFHRRTEKNKRVYMIGNGSS--NIKTEFD 293
Query 314 LTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFI 373
LTEK I+PMGGFPHYGEVR+DFIM+KG +VG +KR +T+RK+L+ +RA E + +KF+
Sbjct 294 LTEKPISPMGGFPHYGEVRNDFIMVKGAVVGARKRVVTLRKSLL---RQRAGEELVIKFV 350
Query 374 DTSSKWGHGRFQTSDEKAKFYGPLKRTAA 402
DTSSK GHGRFQTS EK FYG K A
Sbjct 351 DTSSKIGHGRFQTSAEKKAFYGARKADIA 379
> Hs17466365
Length=251
Score = 257 bits (656), Expect = 4e-68, Method: Compositional matrix adjust.
Identities = 144/303 (47%), Positives = 186/303 (61%), Gaps = 63/303 (20%)
Query 68 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSE 127
MTHIV EV+RPGSK++KKEVVEAVTIVE MV G+VGY+ TP+GL TV+ H+S+
Sbjct 1 MTHIVWEVDRPGSKVNKKEVVEAVTIVE-ITMVVAGIVGYVGTPRGLWTFKTVFTEHISD 59
Query 128 ECRRRFYKNWYKSKKKAFTRYSKKYA-ENNKMQAEID--SIKQHCAVIRAICHTQPSKTP 184
EC+R FYKNW+KSKKKAFT+Y KK+ E+ K Q E D S+K++C
Sbjct 60 ECKRHFYKNWHKSKKKAFTKYCKKWLDEDGKKQLEKDFSSMKKYC--------------- 104
Query 185 TGLRKAHIMEIQVNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVV 244
+KAH+MEIQVN GTVA+K+D+ + E VP++ VF QDEM+DV
Sbjct 105 ---QKAHVMEIQVNRGTVAEKLDWARERLEQQVPVNQVFGQDEMIDV------------- 148
Query 245 SRFGVTRLPRKTHRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSD 304
IGAWHP RV F R GQK Y H T++NKK+Y+IG
Sbjct 149 ---------------------IGAWHPDRVAFSGARTGQKEYHHCTKINKKIYKIGQSYL 187
Query 305 PR-------NGSTDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLV 357
+ N STD DL+ K I P+GGF HYGEV +DF+M+KGC+V KKR +T+ K+L+
Sbjct 188 IKGGKLIKNNASTDYDLSVKSINPLGGFVHYGEVTNDFVMLKGCVVVTKKRVLTLHKSLL 247
Query 358 PRT 360
+T
Sbjct 248 VQT 250
> Hs22055156
Length=222
Score = 218 bits (554), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 113/210 (53%), Positives = 143/210 (68%), Gaps = 8/210 (3%)
Query 197 VNGGTVADKVDFVTKMFESPVPISAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLPRKT 256
VN GTVA+K+D+ K E VP++ VF QDEM+ VIGVTKG G KGV S + T+LP KT
Sbjct 5 VNRGTVAEKLDWAQKRLEQQVPVNQVFGQDEMIKVIGVTKGKGYKGVTSCWHTTKLPHKT 64
Query 257 HRGLRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSD-------PRNGS 309
HRGL KVACIGAWH A V F V GQKGY H TE+NKK+Y+IG G N S
Sbjct 65 HRGLCKVACIGAWHLAHVAFSVSHAGQKGYHHGTEINKKIYKIGQGYPIMDGKLIKNNAS 124
Query 310 TDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIH 369
D DL++K I +GGF H G+V +DF+M+KG +VG KK+ +T+ K+L+ +T +ALE
Sbjct 125 IDYDLSDKSINSLGGFVHCGKVTNDFVMLKGYVVGTKKQVLTLCKSLLVQTKWQALEKTD 184
Query 370 LKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
LKF DT+S +GH FQT +KA F PLK+
Sbjct 185 LKFTDTTSNFGHCHFQTMKKKA-FVRPLKK 213
> 7299422
Length=148
Score = 187 bits (475), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 87/113 (76%), Positives = 98/113 (86%), Gaps = 0/113 (0%)
Query 29 LGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREVERPGSKLHKKEVV 88
+ F P+KR RHRGKVKAFPKDD SKP HLT F+GYKAGMTHIVRE +RPGSK++KKEVV
Sbjct 1 MAFYPKKRSARHRGKVKAFPKDDASKPVHLTCFIGYKAGMTHIVREADRPGSKINKKEVV 60
Query 89 EAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYKNWYKSK 141
EAVT++E+PPM+ VG VGYIETP GLRAL VWA HLSEECRRRFYKNWY S+
Sbjct 61 EAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSEECRRRFYKNWYVSE 113
> 7299421
Length=125
Score = 181 bits (458), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 84/108 (77%), Positives = 94/108 (87%), Gaps = 0/108 (0%)
Query 29 LGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREVERPGSKLHKKEVV 88
+ F P+KR RHRGKVKAFPKDD SKP HLT F+GYKAGMTHIVRE +RPGSK++KKEVV
Sbjct 1 MAFYPKKRSARHRGKVKAFPKDDASKPVHLTCFIGYKAGMTHIVREADRPGSKINKKEVV 60
Query 89 EAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYKN 136
EAVT++E+PPM+ VG VGYIETP GLRAL VWA HLSEECRRRFYKN
Sbjct 61 EAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSEECRRRFYKN 108
> Hs18583607
Length=169
Score = 172 bits (437), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 86/156 (55%), Positives = 112/156 (71%), Gaps = 9/156 (5%)
Query 252 LPRKTHRG-LRKVACIGAWHPARVQFQVPRHGQKGYFHRTEMNKKVYRIGSGSDPRNG-- 308
LPR+ G + + H A ++F V R GQKGY HRTE+NKK+Y+IG G ++G
Sbjct 6 LPRQEAEGPMSGSGELSVRHHA-IKFSVARAGQKGYHHRTEINKKIYKIGQGYLIKDGKL 64
Query 309 -----STDADLTEKRITPMGGFPHYGEVRSDFIMIKGCIVGCKKRPITMRKTLVPRTSRR 363
STD DL++K I P+GGF HYGEV +DF+M+KGC+VG KKR +T+RK+L+ +T RR
Sbjct 65 IKNNASTDYDLSDKSINPLGGFVHYGEVTNDFVMLKGCVVGTKKRVLTLRKSLLVQTKRR 124
Query 364 ALENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
ALE I LKFIDT+SK+GHGRFQT +EK F GPLK+
Sbjct 125 ALEKIDLKFIDTTSKFGHGRFQTMEEKKAFMGPLKK 160
> Hs18596957
Length=133
Score = 111 bits (277), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 53/84 (63%), Positives = 66/84 (78%), Gaps = 3/84 (3%)
Query 99 MVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYKNWYKSKKKAFTRYSKKYA-ENNK 157
M+ VG+VGY+ETPQGLR TV+A H+S+EC+RRFYKNW+KSKKKAFT+Y KK+ EN K
Sbjct 1 MMVVGIVGYVETPQGLRTFKTVFAEHISDECKRRFYKNWHKSKKKAFTKYRKKWQDENGK 60
Query 158 MQAEID--SIKQHCAVIRAICHTQ 179
Q E D S+K++C VI I HTQ
Sbjct 61 KQLEKDFSSMKKYCQVIHVIAHTQ 84
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/38 (57%), Positives = 27/38 (71%), Gaps = 0/38 (0%)
Query 362 RRALENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
RRAL+ LKF DT+SK+GHGR QT +EK G LK+
Sbjct 87 RRALQKTDLKFTDTTSKFGHGRLQTMEEKKASMGRLKK 124
> Hs18564863
Length=103
Score = 103 bits (258), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 49/62 (79%), Positives = 53/62 (85%), Gaps = 0/62 (0%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PRHGSLGF P KR RH GKVK+FPKDDPSKP HLTAF+GYKAGMTHIVR +
Sbjct 1 MSHRKFSTPRHGSLGFLPWKRSSRHCGKVKSFPKDDPSKPVHLTAFLGYKAGMTHIVRAL 60
Query 76 ER 77
E+
Sbjct 61 EK 62
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/37 (70%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 363 RALENIHLKFIDTSSKWGHGRFQTSDEKAKFYGPLKR 399
RALE I LKFIDT SK+GHG FQT +EK F GPLK+
Sbjct 58 RALEKIDLKFIDTPSKFGHGHFQTMEEKKAFMGPLKK 94
> Hs18566516
Length=364
Score = 94.7 bits (234), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 52/119 (43%), Positives = 66/119 (55%), Gaps = 24/119 (20%)
Query 279 PRHGQKGYFHRTEMNKKVYRIGSGSDPR-------NGSTDADLTEKRITPMGGFPHYGEV 331
P QKGY H TE NKK+ IG G + N STD DL++K I P+GGF HY E+
Sbjct 261 PMEIQKGYHHCTENNKKICNIGHGYLTKDEKWIKNNASTDGDLSDKSINPLGGFVHYNEM 320
Query 332 RSDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFIDTSSKWGHGRFQTSDEK 390
+DF+M+KGC+ T R ALE LKFI+T S++G FQT +EK
Sbjct 321 NNDFVMLKGCV-----------------TKRPALEKTDLKFINTVSRFGRDHFQTMEEK 362
Score = 60.5 bits (145), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 28/40 (70%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 28 SLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAG 67
S GF P KR R GKVK+FPKD+PSKP HLTAF+GYKA
Sbjct 188 SRGFLPWKRSSRLHGKVKSFPKDEPSKPFHLTAFLGYKAA 227
> Hs17452839
Length=180
Score = 88.2 bits (217), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 63/178 (35%), Positives = 80/178 (44%), Gaps = 58/178 (32%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PR GSL F P + RH GKVK+F K DPS+ H F+ YKAGMT+I
Sbjct 1 MSHRKFSAPRQGSLSFLPGRHGSRHHGKVKSFSKLDPSQSVHFMVFLVYKAGMTYI---- 56
Query 76 ERPGSKLHKKEVVEAVTIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYK 135
VW E R K
Sbjct 57 --------------------------------------------VW------EVCRPGSK 66
Query 136 NWYKSKKKAFTRYSKKYAENN---KMQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKA 190
NW+KS+KKAFT Y KK+ + + +++ +S+K++C VI I HTQ P RKA
Sbjct 67 NWHKSEKKAFTEYCKKWQDEDGEKQLEKNFNSMKKYCQVICIIAHTQMCLLPL-CRKA 123
> Hs17472282
Length=168
Score = 87.4 bits (215), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 42/60 (70%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 16 MSHRKFERPRHGSLGFCPRKRCRRHRGKVKAFPKDDPSKPPHLTAFMGYKAGMTHIVREV 75
MSHRKF PR G+ GF P+K RH GKVK+FPKDDPSKP HLTA +GYKAGMTHI+ +V
Sbjct 1 MSHRKFSTPRQGAPGFLPQKCSGRHPGKVKSFPKDDPSKPVHLTALLGYKAGMTHILCKV 60
> Hs20472413
Length=132
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 61/127 (48%), Gaps = 22/127 (17%)
Query 282 GQKGYFHRTEMN-------KKVYRIGSGSDPRNGSTDADLTEK--RITPMGGFPHYGEVR 332
G + Y+ R + N K V + + R G+ D DL E+ R+ GG
Sbjct 12 GWQRYYQREDENLELRGPHKPVVKEDPVAKMREGARDPDLFEEMPRVGKAGGH------- 64
Query 333 SDFIMIKGCIVGCKKRPITMRKTLVPRTSRRALENIHLKFIDTSSKWGHGRFQTSDEKAK 392
M K G + ++K + +T R LE I LKFIDT+SK+GHG FQT +EK
Sbjct 65 ----MAKHIHGGPPS--VKLKKLNLHQTKWRVLEKIDLKFIDTTSKFGHGCFQTIEEKKA 118
Query 393 FYGPLKR 399
F PLK+
Sbjct 119 FMEPLKK 125
> Hs6005862
Length=348
Score = 35.0 bits (79), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 8/87 (9%)
Query 220 SAVFEQDEMLDVIGVTKGHGVKGVVSRFGVTRLP-----RKTHRGLRKVACIGAWHPARV 274
+A F + +DV T G G +GV+ R+G P KTH R+ + RV
Sbjct 195 AAHFRPGQYVDVTAKTIGKGFQGVMKRWGFKGQPATHGQTKTH---RRPGAVATGDIGRV 251
Query 275 QFQVPRHGQKGYFHRTEMNKKVYRIGS 301
G+ G +RTE KV+RI +
Sbjct 252 WPGTKMPGKMGNIYRTEYGLKVWRINT 278
> At3g43530
Length=615
Score = 30.8 bits (68), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 38/87 (43%), Gaps = 5/87 (5%)
Query 92 TIVESPPMVCVGLVGYIETPQGLRALSTVWAGHLSEECRRRFYKNWYKSKKKAFTRYSKK 151
T+V++P V G +T L + S + E + +N +KS K T +SKK
Sbjct 331 TVVQAPRKAAVEKKGKGKTAAALTSPSDEGLTEVVNEMKN-LMENGFKSMNKRMTNFSKK 389
Query 152 YAENNK----MQAEIDSIKQHCAVIRA 174
Y E +K M+ I SI+ A
Sbjct 390 YEEQDKRLKLMETAIKSIQSSTGTDDA 416
> CE20896
Length=458
Score = 30.4 bits (67), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query 149 SKKYAENNKMQAEIDSIKQHCAVIRAICHTQPSKTPTGLRKAHIMEIQVNG-------GT 201
SK++ N + + ++QH +I A+C PS T + + + + NG G
Sbjct 276 SKRHPNNADISKTVSLVRQHHGIIHAVCSVAPSGG-TQSKAMYNLTSKTNGVCNIGEDGD 334
Query 202 VADKVDFVTKMFESPVPI 219
+ +D++ ++++P PI
Sbjct 335 FDELMDYI-PIYDAPYPI 351
Lambda K H
0.322 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10210178872
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40