bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0903_orf1
Length=162
Score E
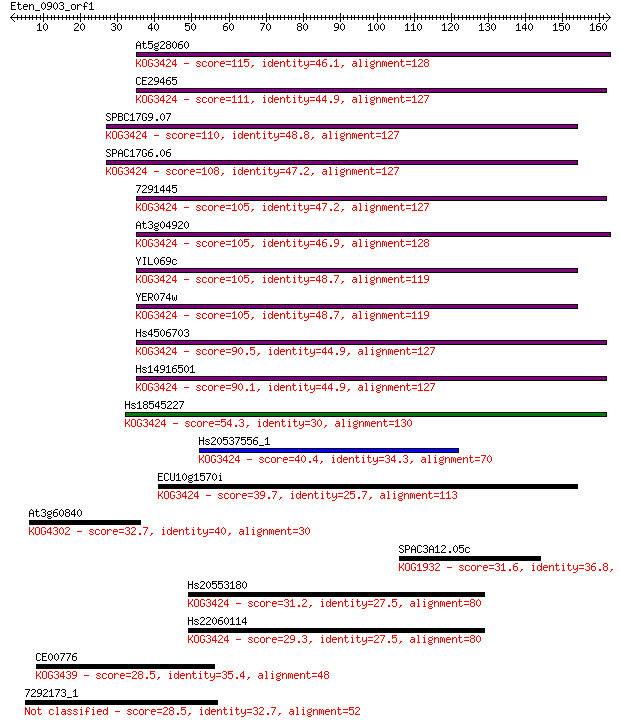
Sequences producing significant alignments: (Bits) Value
At5g28060 115 3e-26
CE29465 111 7e-25
SPBC17G9.07 110 1e-24
SPAC17G6.06 108 4e-24
7291445 105 4e-23
At3g04920 105 4e-23
YIL069c 105 5e-23
YER074w 105 5e-23
Hs4506703 90.5 1e-18
Hs14916501 90.1 2e-18
Hs18545227 54.3 9e-08
Hs20537556_1 40.4 0.002
ECU10g1570i 39.7 0.003
At3g60840 32.7 0.36
SPAC3A12.05c 31.6 0.72
Hs20553180 31.2 0.90
Hs22060114 29.3 3.2
CE00776 28.5 5.3
7292173_1 28.5 5.9
> At5g28060
Length=133
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 84/128 (65%), Gaps = 1/128 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R R +N LL RKQ +DVLH GRANVS+ EL+++LA+ ++V DP + FRT F
Sbjct 7 TIRTRNFMTNRLLARKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFCFKFRTHF 66
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S G LIY+ A++FE ++RL+R G + + +K R+ KE KNR KK+RG +K
Sbjct 67 GGGKSSGYGLIYDTVENAKKFEPKYRLIRNG-LDTKIEKSRKQIKERKNRAKKIRGVKKT 125
Query 155 KTQAGKQK 162
K K+K
Sbjct 126 KAGDTKKK 133
> CE29465
Length=329
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 86/127 (67%), Gaps = 1/127 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ +N LL RKQ+ V+V+H GR V + ++++++A+ +K P ++ F +
Sbjct 6 TIRTRKVLTNKLLYRKQMVVEVIHPGRPTVPKADIREKIAKLYKT-TPDTVIPFGFESKI 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S+G AL+Y+ A++FE ++RLVRMG A K GR+ KE KNR+KKVRGT KA
Sbjct 65 GGGKSKGFALVYDTIDFAKKFEPKYRLVRMGLATKVEKPGRKQRKERKNRQKKVRGTAKA 124
Query 155 KTQAGKQ 161
K AGK+
Sbjct 125 KVSAGKK 131
> SPBC17G9.07
Length=134
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 62/127 (48%), Positives = 83/127 (65%), Gaps = 4/127 (3%)
Query 27 MSEPKSPFTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLV 86
MSE T+R RK +N LLQRKQ+ VD+LH G+AN+S+ E++++LAQ +K D C+
Sbjct 1 MSEA---VTIRTRKFMTNRLLQRKQMVVDILHPGKANLSKNEIREKLAQMYKT-DSECVQ 56
Query 87 LCCFRTAFGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRK 146
RT FGG RS G ALIY+ + ++FE +RLVR+G AE K R+ K+ KNR K
Sbjct 57 AFGLRTHFGGGRSTGFALIYDSTESMKKFEPHYRLVRVGQAEPIQKVARQQRKQRKNRGK 116
Query 147 KVRGTEK 153
KV GT K
Sbjct 117 KVFGTGK 123
> SPAC17G6.06
Length=134
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 82/127 (64%), Gaps = 4/127 (3%)
Query 27 MSEPKSPFTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLV 86
MSE T+R RK +N LLQRKQ+ VD+LH G+AN+S+ +L+++L Q +K D +
Sbjct 1 MSEA---VTIRTRKFMTNRLLQRKQMVVDILHPGKANISKNDLREKLGQMYKT-DASAVQ 56
Query 87 LCCFRTAFGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRK 146
RT +GG R+ G ALIY+D A ++FE +RLVR+G AE K R+ K+ KNR K
Sbjct 57 AFGLRTHYGGGRTTGFALIYDDVEAMKKFEPHYRLVRVGHAEPIQKVARQQRKQRKNRGK 116
Query 147 KVRGTEK 153
KV GT K
Sbjct 117 KVFGTGK 123
> 7291445
Length=131
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 83/127 (65%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LL RKQ+ DVLH G ++V++ E++++LA +KV P + FRT F
Sbjct 7 TIRTRKFMTNRLLARKQMVCDVLHPGLSSVNKTEIREKLAAMYKVT-PDVVFAFGFRTNF 65
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG RS G ALIY+ A++FE ++RL R G E + K+ R+ KE +NR KKVRGT KA
Sbjct 66 GGGRSTGFALIYDTLDFAKKFEPKYRLARHGLFE-QKKQTRKQRKERRNRMKKVRGTAKA 124
Query 155 KTQAGKQ 161
K GK+
Sbjct 125 KIGTGKK 131
> At3g04920
Length=133
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 60/128 (46%), Positives = 87/128 (67%), Gaps = 1/128 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LL RKQ +DVLH GRANVS+ EL+++LA+ ++V DP + + FRT F
Sbjct 7 TIRTRKFMTNRLLSRKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFVFKFRTHF 66
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S G LIY+ +A++FE ++RL+R G + + +K R+ KE KNR KK+RG +K
Sbjct 67 GGGKSSGFGLIYDTVESAKKFEPKYRLIRNG-LDTKIEKSRKQIKERKNRAKKIRGVKKT 125
Query 155 KTQAGKQK 162
K K+K
Sbjct 126 KAGDAKKK 133
> YIL069c
Length=135
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 58/119 (48%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ SNPLL RKQ VDVLH RANVS+ EL+++LA+ +K + + + FRT F
Sbjct 6 TIRTRKVISNPLLARKQFVVDVLHPNRANVSKDELREKLAEVYK-AEKDAVSVFGFRTQF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
GG +S G L+Y A++FE +RLVR G AE K R+ K+ KNR KK+ GT K
Sbjct 65 GGGKSVGFGLVYNSVAEAKKFEPTYRLVRYGLAEKVEKASRQQRKQKKNRDKKIFGTGK 123
> YER074w
Length=135
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 58/119 (48%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ SNPLL RKQ VDVLH RANVS+ EL+++LA+ +K + + + FRT F
Sbjct 6 TIRTRKVISNPLLARKQFVVDVLHPNRANVSKDELREKLAEVYK-AEKDAVSVFGFRTQF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
GG +S G L+Y A++FE +RLVR G AE K R+ K+ KNR KK+ GT K
Sbjct 65 GGGKSVGFGLVYNSVAEAKKFEPTYRLVRYGLAEKVEKASRQQRKQKKNRDKKIFGTGK 123
> Hs4506703
Length=133
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 81/127 (63%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL R G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NVGAGKK 130
> Hs14916501
Length=130
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 81/127 (63%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL R G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NVGAGKK 130
> Hs18545227
Length=110
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 59/130 (45%), Gaps = 25/130 (19%)
Query 32 SPFTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFR 91
T+ R+L +N LLQRKQ+ +D+L+ G+A V + E+ ++L + +K V FR
Sbjct 3 DTVTIWTRELMTNQLLQRKQMVIDILNTGKATVPKTEILEKLVKIYKTTLDVIFVF-GFR 61
Query 92 TAFGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGT 151
T FGG + F L M + K R+ + KK+RG
Sbjct 62 TYFGGGKIAD-----------------FDLQDMACLHEKKKTSRK-------QMKKLRGL 97
Query 152 EKAKTQAGKQ 161
KA AGK+
Sbjct 98 AKANVGAGKK 107
> Hs20537556_1
Length=84
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query 52 LWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSRGQALIYEDFRA 111
+ +DVLH G+A V + E+Q++LA+ +K V FRT GG G +IY+
Sbjct 1 MLIDVLHPGKATVPKTEIQKKLAKMYKTTLDVIFVF-AFRTHAGG----GFGMIYDSLDY 55
Query 112 AQRFERRFRL 121
+ R RL
Sbjct 56 VKAATLRVRL 65
> ECU10g1570i
Length=126
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 57/115 (49%), Gaps = 6/115 (5%)
Query 41 LQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSR 100
L+ N LL RK+L V H G S++E+++++++ ++ +V+ + +G R+
Sbjct 5 LRDNRLLSRKELDVVAYHPGMPIPSKEEIREKISELYQTKKDNVVVM-DLKNRYGTHRTT 63
Query 101 GQALIYEDFRAAQRFERRFRLVRMGAAE--AEPKKGRRATKELKNRRKKVRGTEK 153
+A IY E++ + ++ E P RR K+ + +R K+ GT K
Sbjct 64 LKAKIYSSIEVLSNIEKKHIVAKLTGKEFHVTP---RRVRKDERKKRYKIFGTLK 115
> At3g60840
Length=648
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 6 APQTHKAVRKEAPAPPAAAAAMSEPKSPFT 35
P+T++ ++ P PAA+ AM+E +PFT
Sbjct 593 TPKTNEEKKRAVPTTPAASVAMTEATTPFT 622
> SPAC3A12.05c
Length=1174
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 106 YEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKN 143
Y FR QRF R+ ++ MG + + K+ RA KN
Sbjct 543 YPIFRVVQRFNRKKMIIEMGIDQVQTKEAPRAPMSDKN 580
> Hs20553180
Length=327
Score = 31.2 bits (69), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 16/80 (20%)
Query 49 RKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSRGQALIYED 108
R+Q+ +DVLH +A + ++Q++LA+ K T FG IY+
Sbjct 253 RQQMVIDVLHPRKATAPKTDIQEKLAEMDKTTQD--------TTGFGT--------IYKS 296
Query 109 FRAAQRFERRFRLVRMGAAE 128
A++ E + RL R E
Sbjct 297 SDYAKKNEPKHRLARHSPCE 316
> Hs22060114
Length=285
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 16/80 (20%)
Query 49 RKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSRGQALIYED 108
R+Q+ +DVLH +A + ++Q++LA+ K T FG IY+
Sbjct 211 RQQMVIDVLHPRKATAPKTDIQEKLAEMDKTTQD--------TTGFGT--------IYKS 254
Query 109 FRAAQRFERRFRLVRMGAAE 128
A++ E + RL R E
Sbjct 255 SDYAKKNEPKHRLARHSPCE 274
> CE00776
Length=118
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 8 QTHKAVRKEAPAPPAAAAAMSEPKSP-FTLRLRKLQSNPLLQRKQLWVD 55
+T A P AAA+ PKS T+RLR + P+L+ K++ V+
Sbjct 2 ETETATTPTGNTEPTAAASAEPPKSDKVTVRLRNIADAPVLKNKKMVVN 50
> 7292173_1
Length=226
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 5 AAPQTHKAVRKEAPAPPAAAAAMSEPKSPFTLRLRKLQSNPLLQRKQLWVDV 56
AAP K V K P PP + + EP+ F + K+ S+P++ + VD+
Sbjct 149 AAPVLPKTVSKTVPTPPPKIS-LKEPRRGFEIPSPKVPSSPVVSGRNTPVDI 199
Lambda K H
0.320 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40