bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0890_orf4
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
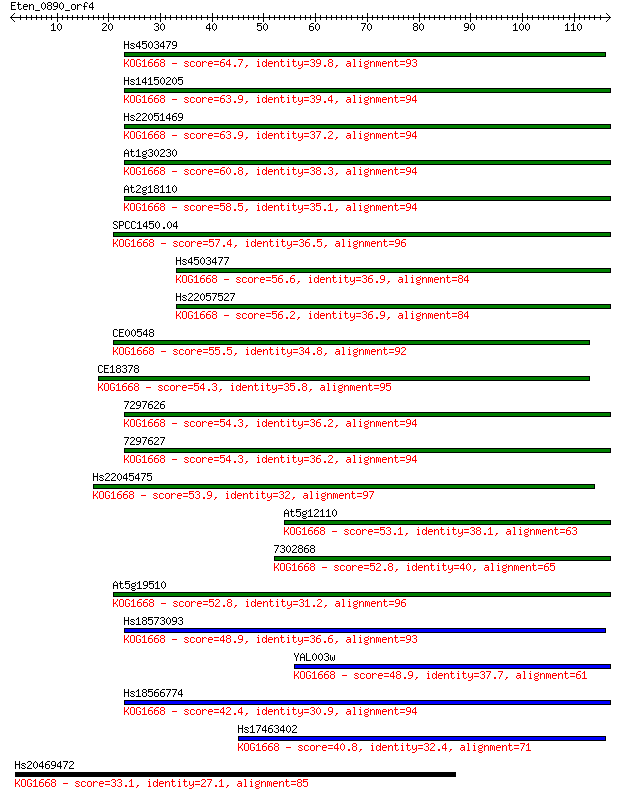
Hs4503479 64.7 4e-11
Hs14150205 63.9 7e-11
Hs22051469 63.9 7e-11
At1g30230 60.8 6e-10
At2g18110 58.5 3e-09
SPCC1450.04 57.4 6e-09
Hs4503477 56.6 1e-08
Hs22057527 56.2 1e-08
CE00548 55.5 3e-08
CE18378 54.3 5e-08
7297626 54.3 6e-08
7297627 54.3 6e-08
Hs22045475 53.9 7e-08
At5g12110 53.1 1e-07
7302868 52.8 1e-07
At5g19510 52.8 2e-07
Hs18573093 48.9 2e-06
YAL003w 48.9 2e-06
Hs18566774 42.4 2e-04
Hs17463402 40.8 6e-04
Hs20469472 33.1 0.12
> Hs4503479
Length=281
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 60/97 (61%), Gaps = 5/97 (5%)
Query 23 LFGDD----DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLT 78
LFG D D A Q +E+ +Q +++ KK +V KSS+++++KP + E D+ ++
Sbjct 159 LFGSDNEEEDKEAAQLREERLRQYAEKKAKKPALVAKSSILLDVKPWDDETDMAQLEACV 218
Query 79 KEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVV 115
+ I L+GL WG S K VPV +G+ KLQ+ C + D+ V
Sbjct 219 RSIQLDGLVWGAS-KLVPVGYGIRKLQIQCVVEDDKV 254
> Hs14150205
Length=647
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 59/98 (60%), Gaps = 5/98 (5%)
Query 23 LFGDD----DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLT 78
LFG D D A Q +E+ +Q +++ KK +V KSS+++++KP + E D+ ++
Sbjct 525 LFGSDNEEEDKEAAQLREERLRQYAEKKAKKPALVAKSSILLDVKPWDDETDMAQLEACV 584
Query 79 KEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+ I L+GL WG S K VPV +G+ KLQ+ C + D V
Sbjct 585 RSIQLDGLVWGAS-KLVPVGYGIRKLQIQCLVEDAKVG 621
> Hs22051469
Length=256
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 60/97 (61%), Gaps = 4/97 (4%)
Query 23 LFGDDDP---AAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTK 79
LFG D+ QL+E++ +Q ++ KK +V KSS+++++KP + E D+ ++ +
Sbjct 135 LFGSDNEEEDKEAAQLREERLRQYAEKAKKPALVAKSSILLDVKPWDDETDMAQLEACVR 194
Query 80 EIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
I L+GL WG S K +PV +G+ KLQ+ C + D+ V
Sbjct 195 SIELDGLVWGTS-KLMPVGYGIRKLQIQCVVEDDKVG 230
> At1g30230
Length=242
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 59/94 (62%), Gaps = 3/94 (3%)
Query 23 LFGDDDPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTKEIV 82
LFG++ + +E+ + KKK+ KSS++I+IKP + E D+ ++ K I
Sbjct 112 LFGEETEEEKKAAEERAASVKASTKKKES--GKSSVLIDIKPWDDETDMKKLEEAVKSIQ 169
Query 83 LEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+EGL WG S K VPV +G+ KLQ+ C+I+D++V+
Sbjct 170 MEGLFWGAS-KLVPVGYGIKKLQILCTIVDDLVS 202
> At2g18110
Length=231
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 58/94 (61%), Gaps = 3/94 (3%)
Query 23 LFGDDDPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTKEIV 82
LFG++ + +E+ + KKK+ KSS++++IKP + E D+ ++ + I
Sbjct 112 LFGEETEEEKKAAEERAASVKASTKKKES--GKSSVLMDIKPWDDETDMKKLEEAVRSIQ 169
Query 83 LEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+EGL WG S K VPV +G+ KL + C+I+D++V+
Sbjct 170 MEGLFWGAS-KLVPVGYGIKKLHIMCTIVDDLVS 202
> SPCC1450.04
Length=214
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 59/99 (59%), Gaps = 4/99 (4%)
Query 21 IDLFGDD---DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARL 77
IDLFG D DP A + E+ + +++ K + V+KS + +++KP + E +DE+ +
Sbjct 91 IDLFGSDEEEDPEAERIKAERVAEYNKKKAAKPKAVHKSLVTLDVKPWDDETPMDELEKA 150
Query 78 TKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+ I ++GL WG S K VPV FG+ K Q+ + D+ V+
Sbjct 151 VRSIQMDGLVWGLS-KLVPVGFGVNKFQINLVVEDDKVS 188
> Hs4503477
Length=225
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 55/85 (64%), Gaps = 2/85 (2%)
Query 33 QQLKEKQQQQQQQQKKKK-EVVNKSSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGES 91
++L+E++ Q + +K KK +V KSS+++++KP + E D+ ++ + I +GL WG S
Sbjct 116 KRLREERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEECVRSIQADGLVWGSS 175
Query 92 VKKVPVAFGLYKLQVCCSILDEVVN 116
K VPV +G+ KLQ+ C + D+ V
Sbjct 176 -KLVPVGYGIKKLQIQCVVEDDKVG 199
> Hs22057527
Length=225
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 55/85 (64%), Gaps = 2/85 (2%)
Query 33 QQLKEKQQQQQQQQKKKK-EVVNKSSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGES 91
++L+E++ Q + +K KK +V KSS+++++KP + E D+ ++ + I +GL WG S
Sbjct 116 KRLREERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEERVRSIQADGLVWGSS 175
Query 92 VKKVPVAFGLYKLQVCCSILDEVVN 116
K VPV +G+ KLQ+ C + D+ V
Sbjct 176 -KLVPVGYGIKKLQIQCVVEDDKVG 199
> CE00548
Length=213
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 56/96 (58%), Gaps = 5/96 (5%)
Query 21 IDLFGDDD----PAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVAR 76
DLFG DD + ++E+ +++ KK + KSS+++++KP + E DL E+ +
Sbjct 88 FDLFGSDDEEEDAEKAKIVEERLAAYAEKKAKKAGPIAKSSVILDVKPWDDETDLGEMEK 147
Query 77 LTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILD 112
L + I ++GL WG K +P+ +G+ KLQ+ I D
Sbjct 148 LVRSIEMDGLVWG-GAKLIPIGYGIKKLQIITVIED 182
> CE18378
Length=285
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 58/99 (58%), Gaps = 5/99 (5%)
Query 18 DDDIDLFGDDDPAAVQQLKEKQQQQQQQQKKKKEV----VNKSSLVIEIKPANAEGDLDE 73
DDD DLFG +D ++ K+ +++ KK + KSS+++++KP + E DL E
Sbjct 157 DDDFDLFGSEDEEEDEEKKKVVEERLAAYAAKKATKAGPIAKSSVILDVKPWDDETDLGE 216
Query 74 VARLTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILD 112
+ +L + I ++GL WG K +P+ +G+ KLQ+ I D
Sbjct 217 MEKLVRSIEMDGLVWG-GAKLIPIGYGIKKLQIITVIED 254
> 7297626
Length=256
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 58/98 (59%), Gaps = 5/98 (5%)
Query 23 LFGDD----DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLT 78
LFG D D A + +E+ ++ KK +++ KS++++++KP + E DL +
Sbjct 134 LFGSDSEEEDGEAARIREERLAAYAAKKAKKVQIIAKSNIILDVKPWDDETDLKVMETEI 193
Query 79 KEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
++I +GL WG S K VPVAFG+ KL + C + D+ V+
Sbjct 194 RKITQDGLLWGAS-KFVPVAFGIQKLSISCVVEDDKVS 230
> 7297627
Length=229
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 58/98 (59%), Gaps = 5/98 (5%)
Query 23 LFGDD----DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLT 78
LFG D D A + +E+ ++ KK +++ KS++++++KP + E DL +
Sbjct 107 LFGSDSEEEDGEAARIREERLAAYAAKKAKKVQIIAKSNIILDVKPWDDETDLKVMETEI 166
Query 79 KEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
++I +GL WG S K VPVAFG+ KL + C + D+ V+
Sbjct 167 RKITQDGLLWGAS-KFVPVAFGIQKLSISCVVEDDKVS 203
> Hs22045475
Length=336
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 55/97 (56%), Gaps = 17/97 (17%)
Query 17 DDDDIDLFGDDDPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVAR 76
+DD++DLFG D ++ K+ +V KSS+++++KP + E D+ ++
Sbjct 80 EDDNMDLFGSDG----------------EEDKEAALVVKSSILLDVKPWDNETDMAQLEA 123
Query 77 LTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDE 113
+ I L+ L WG S K VP+ +G+ KLQ+ C + D+
Sbjct 124 CARSIQLDRLVWGAS-KLVPMGYGIQKLQIQCVMEDD 159
> At5g12110
Length=228
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 43/63 (68%), Gaps = 1/63 (1%)
Query 54 NKSSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDE 113
KSS+++E+KP + E D+ ++ + + + GLTWG S K VPV +G+ KL + +I+D+
Sbjct 138 GKSSVLLEVKPWDDETDMKKLEEAVRSVQMPGLTWGAS-KLVPVGYGIKKLTIMMTIVDD 196
Query 114 VVN 116
+V+
Sbjct 197 LVS 199
> 7302868
Length=222
Score = 52.8 bits (125), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 44/65 (67%), Gaps = 1/65 (1%)
Query 52 VVNKSSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSIL 111
++ KSS+++++KP + E D+ E+ + I ++GL WG S K VPV +G+ KLQ+ C I
Sbjct 133 LIAKSSVLLDVKPWDDETDMKEMENNVRTIEMDGLLWGAS-KLVPVGYGINKLQIMCVIE 191
Query 112 DEVVN 116
D+ V+
Sbjct 192 DDKVS 196
> At5g19510
Length=224
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 60/96 (62%), Gaps = 3/96 (3%)
Query 21 IDLFGDDDPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTKE 80
+DLFGD+ + +E++ ++ +K K+ KSS+++++KP + E D+ ++ +
Sbjct 103 MDLFGDETEEEKKAAEEREAAKKDTKKPKES--GKSSVLMDVKPWDDETDMKKLEEAVRG 160
Query 81 IVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+ + GL WG S K VPV +G+ KL + +I+D++V+
Sbjct 161 VEMPGLFWGAS-KLVPVGYGIKKLTIMFTIVDDLVS 195
> Hs18573093
Length=274
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 54/97 (55%), Gaps = 11/97 (11%)
Query 23 LFGDD----DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLT 78
LFG D D A Q +E+ QQ +++ KK E+V KSS++++ KP + E D+ ++
Sbjct 159 LFGSDNEEEDKKAAQLQEERLQQYAEKKAKKPELVAKSSILLDFKPWDDETDMAQLEACV 218
Query 79 KEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVV 115
I LE L VP+ +G+ KLQ+ C + D+ V
Sbjct 219 CSIQLEELL-------VPMGYGIRKLQIQCVVEDDKV 248
> YAL003w
Length=206
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 40/61 (65%), Gaps = 1/61 (1%)
Query 56 SSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGESVKKVPVAFGLYKLQVCCSILDEVV 115
S + +++KP + E +L+E+ K I +EGLTWG + +P+ FG+ KLQ+ C + D+ V
Sbjct 121 SIVTLDVKPWDDETNLEEMVANVKAIEMEGLTWGAH-QFIPIGFGIKKLQINCVVEDDKV 179
Query 116 N 116
+
Sbjct 180 S 180
> Hs18566774
Length=180
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 45/94 (47%), Gaps = 15/94 (15%)
Query 23 LFGDDDPAAVQQLKEKQQQQQQQQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTKEIV 82
LFG D ++ K +++ Q + KK AN E D+ ++ + I
Sbjct 76 LFGSDYEEESEEAKRLREEHLAQYESKK--------------ANDETDMAKLEECVRSIQ 121
Query 83 LEGLTWGESVKKVPVAFGLYKLQVCCSILDEVVN 116
+GL WG S K VPV +G+ KLQ+ C + D+ V
Sbjct 122 ADGLVWGSS-KLVPVGYGIKKLQIQCVVEDDKVG 154
> Hs17463402
Length=185
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 45 QQKKKKEVVNKSSLVIEIKPANAEGDLDEVARLTKEIVLEGLTWGESVKKVPVAFGLYKL 104
++ K +V KSS++++IKP + + D+ ++ + I L+GL G S K V +G+ K+
Sbjct 89 KKAKNPSLVAKSSILLDIKPWDNKTDMTQLEACVRSIQLDGLVSGAS-KLASVGYGIRKM 147
Query 105 QVCCSILDEVV 115
Q+ C + D+ V
Sbjct 148 QIQCVVEDDKV 158
> Hs20469472
Length=132
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 3/88 (3%)
Query 2 AGQQQQQQQQQQQQQDDDDIDLFGDD---DPAAVQQLKEKQQQQQQQQKKKKEVVNKSSL 58
A + + +DDDDIDL G D + A ++L E+ Q + +K KK +
Sbjct 44 ANVEDTTGSRAANSKDDDDIDLLGSDKEEEGEAAKRLMEEWLAQNESKKAKKHALVAIFH 103
Query 59 VIEIKPANAEGDLDEVARLTKEIVLEGL 86
++KP + D+ ++ + I +GL
Sbjct 104 HTDVKPWDDVADMAKLEECVRSIQADGL 131
Lambda K H
0.310 0.128 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40