bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0885_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
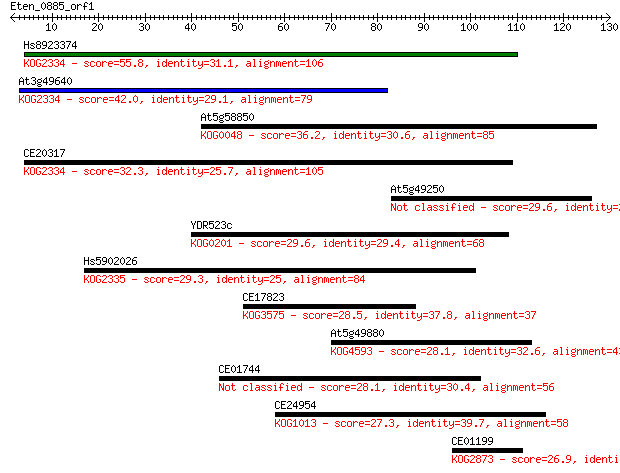
Hs8923374 55.8 2e-08
At3g49640 42.0 3e-04
At5g58850 36.2 0.014
CE20317 32.3 0.25
At5g49250 29.6 1.5
YDR523c 29.6 1.6
Hs5902026 29.3 2.0
CE17823 28.5 3.5
At5g49880 28.1 3.7
CE01744 28.1 4.4
CE24954 27.3 7.3
CE01199 26.9 9.5
> Hs8923374
Length=493
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 54/106 (50%), Gaps = 10/106 (9%)
Query 4 DATRFQDSYHIGNLMFARGAMWDPSLFAQKKVVTTEGTAACSFERTAVLQDYIKRAVLCG 63
D F+ + ++M AR AMW+PS+F ++ + E V+Q YI+ AV
Sbjct 226 DIEDFRQATAASSVMVARAAMWNPSIFLKEGLRPLE----------EVMQKYIRYAVQYD 275
Query 64 SPYQSIKFTLQEMTARDSDKKLKLDLVAANSTAKLCDVFGLGDFYK 109
+ Y + K+ L +M + L AA S+ ++C+ FGLG FY+
Sbjct 276 NHYTNTKYCLCQMLREQLESPQGRLLHAAQSSREICEAFGLGAFYE 321
> At3g49640
Length=519
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 43/79 (54%), Gaps = 10/79 (12%)
Query 3 EDATRFQDSYHIGNLMFARGAMWDPSLFAQKKVVTTEGTAACSFERTAVLQDYIKRAVLC 62
+D +R + + ++M ARGAMW+ S+F+ K E V + Y+++++L
Sbjct 431 DDFSRIKTATGAASVMVARGAMWNASIFSPKGKSHWED----------VKKKYLRKSILW 480
Query 63 GSPYQSIKFTLQEMTARDS 81
+ +S K+T++EM A S
Sbjct 481 NNDVKSTKYTIKEMIAHHS 499
> At5g58850
Length=449
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 43/85 (50%), Gaps = 13/85 (15%)
Query 42 AACSFERTAVLQDYIKRAVLCGSPYQSIKFTLQEMTARDSDKKLKLDLVAANSTAKLCDV 101
A+ S +R +LQDYIK SI+ + +D+D+K + ++ ST L +
Sbjct 243 ASPSAKRPCILQDYIK----------SIE---RNNINKDNDEKKNENTISVISTPNLDQI 289
Query 102 FGLGDFYKSIEHVPYANTLNYYKHI 126
+ GD SI PY L+Y+++I
Sbjct 290 YSDGDSASSILGGPYDEELDYFQNI 314
> CE20317
Length=436
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 48/107 (44%), Gaps = 13/107 (12%)
Query 4 DATRFQDSYHIGNLMFARGAMWDPSLFAQKKVVTTEGTAACSFERTAVLQDYIKRAVLCG 63
D ++Q + M AR A+ PS+F + EG ++ ++++++ A
Sbjct 220 DFEKYQLLTETSSTMIARKALSTPSIFRR------EGC----LDKYEDIRNFLELACQYD 269
Query 64 SPYQSIKFTLQEMTARDS--DKKLKLDLVAANSTAKLCDVFGLGDFY 108
Y K+ +Q + D D + K VAA S ++C FG+ D Y
Sbjct 270 ESYTMTKYVVQRILGADQEYDPRGKA-TVAAGSVLQICKAFGMEDVY 315
> At5g49250
Length=200
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 29/43 (67%), Gaps = 1/43 (2%)
Query 83 KKLKLDLVAANSTAKLCDVFGLGDFYKSIEHVPYANTLNYYKH 125
K+LK+ ++++ + + C + G+G F+ + H+P +++L +KH
Sbjct 52 KELKIVVLSSLNRSYCCGICGIGLFF-FVAHLPLSSSLFVFKH 93
> YDR523c
Length=490
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 32/72 (44%), Gaps = 12/72 (16%)
Query 40 GTAACSFERTAVLQDYIKRAVLCGSPYQSIKFTLQEMTA----RDSDKKLKLDLVAANST 95
G +CS D +KR+ + G P + + F + E+T +K+ D+ AAN
Sbjct 97 GGGSCS--------DLLKRSYVNGLPEEKVSFIIHEVTLGLKYLHEQRKIHRDIKAANIL 148
Query 96 AKLCDVFGLGDF 107
+ LGDF
Sbjct 149 LNEEGMVKLGDF 160
> Hs5902026
Length=307
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 40/86 (46%), Gaps = 15/86 (17%)
Query 17 LMFARGAMWDPSLFAQKKVVTTEGTAACSFERTAV--LQDYIKRAVLCGSPYQSIKFTLQ 74
+M ARG + +P++FA +E T + + D++ A+ G+PY L
Sbjct 227 VMVARGLLANPAMFA-------------GYEETPLKCIWDWVDIALELGTPYMCFHQHLM 273
Query 75 EMTARDSDKKLKLDLVAANSTAKLCD 100
M + + ++ K A +ST+ + D
Sbjct 274 YMMEKITSRQEKRVFNALSSTSAIID 299
> CE17823
Length=407
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 6/43 (13%)
Query 51 VLQDYIKRAVLCGSPYQ------SIKFTLQEMTARDSDKKLKL 87
V +D +KR V PYQ + +L+E+ +R+ D+K+K+
Sbjct 141 VARDILKRDVSTKIPYQFHNSLERLALSLEEIRSREPDEKIKM 183
> At5g49880
Length=726
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 70 KFTLQEMTARDSDKKLKLDLVAANSTAKLCDVFGLGDFYKSIE 112
+FTLQ + A+ D+KL+ + + N++ + GD K IE
Sbjct 657 RFTLQSIYAQSDDEKLEFEYESGNTSILNNEYASQGDIAKQIE 699
> CE01744
Length=4385
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Query 46 FERTAVLQDYIKRAVLCGSPYQSIKFTLQEMTARDSDKKLKLDLVAANSTAKLCDV 101
E+T V ++ I R ++ ++IK +E A D KLK++L+ N ++CD+
Sbjct 4297 LEKTFVREE-IDRTIIGARKGENIKKKFEE--AGDEKVKLKVELIGGNLPNEICDI 4349
> CE24954
Length=1069
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 29/62 (46%), Gaps = 8/62 (12%)
Query 58 RAVLCGSPYQ----SIKFTLQEMTARDSDKKLKLDLVAANSTAKLCDVFGLGDFYKSIEH 113
R +LC S Q SI TL T +DKKLK+ L+ A + K D G D Y
Sbjct 779 RCLLCFSRKQRSLGSITLTL---TYHSADKKLKMHLIRAKNL-KAMDSNGFSDPYVKFHL 834
Query 114 VP 115
+P
Sbjct 835 LP 836
> CE01199
Length=190
Score = 26.9 bits (58), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 10/15 (66%), Positives = 12/15 (80%), Gaps = 0/15 (0%)
Query 96 AKLCDVFGLGDFYKS 110
AKLC+ FGLGD+ S
Sbjct 34 AKLCEAFGLGDYMSS 48
Lambda K H
0.320 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40