bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0863_orf3
Length=189
Score E
Sequences producing significant alignments: (Bits) Value
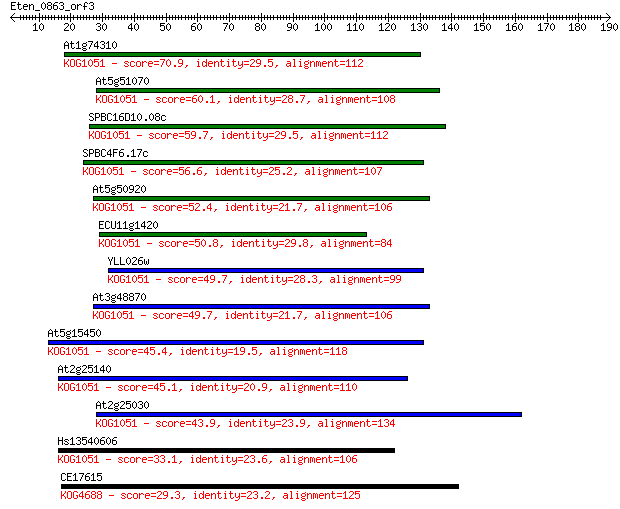
At1g74310 70.9 1e-12
At5g51070 60.1 3e-09
SPBC16D10.08c 59.7 3e-09
SPBC4F6.17c 56.6 3e-08
At5g50920 52.4 6e-07
ECU11g1420 50.8 1e-06
YLL026w 49.7 3e-06
At3g48870 49.7 4e-06
At5g15450 45.4 6e-05
At2g25140 45.1 1e-04
At2g25030 43.9 2e-04
Hs13540606 33.1 0.32
CE17615 29.3 4.8
> At1g74310
Length=911
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 67/112 (59%), Gaps = 0/112 (0%)
Query 18 GNSPKEVLAKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKEL 77
G + K + R VM+EV +F+P+++ R+ +IV+F+PLS +R V L + L
Sbjct 729 GLTGKVTMEVARDCVMREVRKHFRPELLNRLDEIVVFDPLSHDQLRKVARLQMKDVAVRL 788
Query 78 KKRGITLVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKL 129
+RG+ L V+D+A +IL ++ +G R + +++EK V +++ +++ ++
Sbjct 789 AERGVALAVTDAALDYILAESYDPVYGARPIRRWMEKKVVTELSKMVVREEI 840
> At5g51070
Length=945
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 57/108 (52%), Gaps = 0/108 (0%)
Query 28 MRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGITLVVS 87
M+ V++E+ YF+P+++ R+ +IVIF L + M +L+LM L L G+ L VS
Sbjct 808 MKALVVEELKNYFRPELLNRIDEIVIFRQLEKAQMMEILNLMLQDLKSRLVALGVGLEVS 867
Query 88 DSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRGHKA 135
+ K I ++ + +G R + + + + V ++ L+ K G A
Sbjct 868 EPVKELICKQGYDPAYGARPLRRTVTEIVEDPLSEAFLAGSFKPGDTA 915
> SPBC16D10.08c
Length=905
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 60/114 (52%), Gaps = 2/114 (1%)
Query 26 AKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKK--RGIT 83
+ R+ VM + G+F+P+ + R++ IVIF L +R +++ + K L+ R I
Sbjct 754 STTREMVMNSIRGFFRPEFLNRISSIVIFNRLRRVDIRNIVENRILEVQKRLQSNHRSIK 813
Query 84 LVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRGHKAEL 137
+ VSD AK + +S +G R + + I+ V +A LIL+ +L+ A +
Sbjct 814 IEVSDEAKDLLGSAGYSPAYGARPLNRVIQNQVLNPMAVLILNGQLRDKETAHV 867
> SPBC4F6.17c
Length=803
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 58/107 (54%), Gaps = 0/107 (0%)
Query 24 VLAKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGIT 83
V K R VM V Y+ P+ + R+ ++F LS+ + ++++ + + L R I
Sbjct 667 VTPKSRDAVMDVVQKYYPPEFLNRIDDQIVFNKLSEKNLEDIVNVRLDEVQQRLNDRRII 726
Query 84 LVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLK 130
L V+++A+ ++ E+ +S +G R + + I+K + +A I+ ++K
Sbjct 727 LTVTEAARKWLAEKGYSPAYGARPLNRLIQKRILNTMAMKIIQGEIK 773
> At5g50920
Length=929
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/106 (21%), Positives = 62/106 (58%), Gaps = 0/106 (0%)
Query 27 KMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGITLVV 86
+++ V +E+ YF+P+ + R+ ++++F L+ ++ + D++ + + LKK+ I L V
Sbjct 787 RIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADILLKEVFERLKKKEIELQV 846
Query 87 SDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRG 132
++ K +++ ++ +G R + + I + + +A +L+ ++K G
Sbjct 847 TERFKERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEG 892
> ECU11g1420
Length=851
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 47/84 (55%), Gaps = 0/84 (0%)
Query 29 RQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGITLVVSD 88
R+K+ + VL F + R+ +++ F L M +L+ +L K+L+ RGIT VSD
Sbjct 721 RKKIEEIVLNRFGAPFVNRIDEVLYFNNLDRENMHRILEYQMGYLEKKLRGRGITFKVSD 780
Query 89 SAKGFILERAWSHKFGGRRMAKYI 112
+ K ++ + S +G R++ ++I
Sbjct 781 AVKEDMVTKVLSSIYGARQLRRFI 804
> YLL026w
Length=908
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 53/101 (52%), Gaps = 2/101 (1%)
Query 32 VMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTK--ELKKRGITLVVSDS 89
VM V +F+P+ + R++ IVIF LS A+ ++D+ + + E + L ++
Sbjct 750 VMGAVRQHFRPEFLNRISSIVIFNKLSRKAIHKIVDIRLKEIEERFEQNDKHYKLNLTQE 809
Query 90 AKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLK 130
AK F+ + +S G R + + I+ + K+A IL ++K
Sbjct 810 AKDFLAKYGYSDDMGARPLNRLIQNEILNKLALRILKNEIK 850
> At3g48870
Length=952
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/106 (21%), Positives = 58/106 (54%), Gaps = 0/106 (0%)
Query 27 KMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGITLVV 86
+++ V +E+ YF+P+ + R+ ++++F L+ ++ + D+M + L+ + I L V
Sbjct 808 RIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVVARLEVKEIELQV 867
Query 87 SDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRG 132
++ K +++ + +G R + + I + + +A +LS +K G
Sbjct 868 TERFKERVVDEGFDPSYGARPLRRAIMRLLEDSMAEKMLSRDIKEG 913
> At5g15450
Length=968
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/122 (18%), Positives = 59/122 (48%), Gaps = 4/122 (3%)
Query 13 AQWNLGNSPKEV----LAKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDL 68
+Q+ L N+ + ++++VM F+P+ + R+ + ++F+PL + ++ L
Sbjct 802 SQFILNNTDDDANELSYETIKERVMNAARSIFRPEFMNRVDEYIVFKPLDREQINRIVRL 861
Query 69 MFTHLTKELKKRGITLVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAK 128
+ K + R + + ++D+A + + +G R + + I++ + ++A IL
Sbjct 862 QLARVQKRIADRKMKINITDAAVDLLGSLGYDPNYGARPVKRVIQQNIENELAKGILRGD 921
Query 129 LK 130
K
Sbjct 922 FK 923
> At2g25140
Length=874
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/110 (20%), Positives = 58/110 (52%), Gaps = 0/110 (0%)
Query 16 NLGNSPKEVLAKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTK 75
N +S + V M+++V++ F+P+ + R+ + ++F+PL + + +++L +
Sbjct 726 NNEDSKEAVYEIMKRQVVELARQNFRPEFMNRIDEYIVFQPLDSNEISKIVELQMRRVKN 785
Query 76 ELKKRGITLVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLIL 125
L+++ I L + A + + + +G R + + I++ V ++A IL
Sbjct 786 SLEQKKIKLQYTKEAVDLLAQLGFDPNYGARPVKRVIQQMVENEIAVGIL 835
> At2g25030
Length=265
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/137 (23%), Positives = 69/137 (50%), Gaps = 10/137 (7%)
Query 28 MRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKELKKRGITLVVS 87
M+Q+V++ F+P+ + R+ + ++ +PL+ S + +++L + K L++ I L +
Sbjct 133 MKQQVVELARKTFRPKFMNRIDEYIVSQPLNSSEISKIVELQMRQVKKRLEQNKINLEYT 192
Query 88 DSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRGHKAE---LKRAPNQP 144
A + + + G R + + IEK V ++ + K+ +G AE + +QP
Sbjct 193 KEAVDLLAQLGFDPNNGARPVKQMIEKLVKKEI-----TLKVLKGDFAEDGTILIDADQP 247
Query 145 NQLNLIICEEDENGGCV 161
N N ++ ++ EN V
Sbjct 248 N--NKLVIKKMENNAHV 262
> Hs13540606
Length=707
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 25/112 (22%), Positives = 55/112 (49%), Gaps = 6/112 (5%)
Query 16 NLGN---SPKEVLAK-MRQKVMQEVL--GYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLM 69
NLG+ S K ++K ++ V++ +L + + + +GR+ +IV F P S + +++
Sbjct 524 NLGDVQISDKITISKNFKENVIRPILKAHFRRDEFLGRINEIVYFLPFCHSELIQLVNKE 583
Query 70 FTHLTKELKKRGITLVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVA 121
K K+R ++ D +L ++ +G R + +E+ V ++A
Sbjct 584 LNFWAKRAKQRHNITLLWDREVADVLVDGYNVHYGARSIKHEVERRVVNQLA 635
> CE17615
Length=451
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 55/125 (44%), Gaps = 10/125 (8%)
Query 17 LGNSPKEVLAKMRQKVMQEVLGYFKPQVIGRMTKIVIFEPLSDSAMRGVLDLMFTHLTKE 76
L E++ +K Q+V G F ++ R + +F +D A++ + + HL +
Sbjct 240 LDTESHEIIEDFSKKPRQQVSGEFLNHLL-RGCYLALFNGAADLALQAIDAI---HLRAQ 295
Query 77 LKKRGITLVVSDSAKGFILERAWSHKFGGRRMAKYIEKYVTAKVAPLILSAKLKRGHKAE 136
+ TL+V +S + +L+ WS + G M + K ++ +A LK
Sbjct 296 YEMLPDTLLVVNSLRNSLLDNMWSKEKRGELMWTRPQHKDQTKRRDMVTNASLK------ 349
Query 137 LKRAP 141
+KR P
Sbjct 350 MKRMP 354
Lambda K H
0.318 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3130490202
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40